The coronavirus disease 2019 (COVID-19) pandemic, caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pathogen, continues to wreak havoc across the globe. Nearly a year on since the virus was first detected in Wuhan, China, in late 2019, over 61.1 million people have been infected across 191 countries, and 1.43 million have lost their lives.
Even as researchers close in on a COVID-19 vaccine, the SARS-CoV-2 virus has undergone many changes in its genetic sequences. This has been observed particularly in the virus’s spike protein (or S protein), which it uses to latch on the human host cell’s angiotensin-converting enzyme 2 (ACE2) receptor to instigate infection.
_-_1200_width.jpg)
Novel Coronavirus SARS-CoV-2 Spike Protein. 3D print of a spike protein of SARS-CoV-2—also known as 2019-nCoV, the virus that causes COVID-19—in front of a 3D print of a SARS-CoV-2 virus particle. The spike protein (foreground) enables the virus to enter and infect human cells. On the virus model, the virus surface (blue) is covered with spike proteins (red) that enable the virus to enter and infect human cells. Image Credit: NIAID / Flickr
The spike protein has thus been the major antigenic target for most antivirals and vaccines. A new preprint published on the bioRxiv* server in November 2020 describes the seven different clades into which the virus has diversified. The important haplotypes (the set of genetic determinants in a chromosome) are also explored. This is relevant in terms of assessing the efficacy of vaccines and the response to prophylactic and therapeutic antivirals, as well as the possibility of reinfection by other clades.
The clades circulating most widely at the start of the pandemic were L, O, S and V; these have been called the founding clades. However, during the course of the pandemic, the D614G mutation emerged in the G clade, in the spike protein, and soon attained a high prevalence. This was followed by the emergence of the GR clade, which soon became dominant in every region where it was introduced. The GH reached its highest frequency at 30% in May 2020, but has subsequently decreased.
The researchers emphasize that these three clades have been suggested to have higher transmissibility, though have not caused a more severe disease.
Only a single study has shown the rate of nucleotide evolution in the spike region of the genome in the first four months of the pandemic, but this did not take account of differences in the seven clades. Others have reported this rate for the whole genome, but this is slower than the mutation capacity of the spike region alone.
The researchers examined 2,100 sequences which represented all seven clades of the virus, to identify the patterns in which their genes had changed, and the rate of change of the nucleotides at the genomic region of the spike.
The scientists found that the various patterns of gene variants that are inherited together, called haplotypes, occurred in the shape of a star. In other words, all of the haplotype networks had a common ancestor, from which many haplotypes diverged with a difference of only a few nucleotides between them.
They found almost 480 haplotypes in these clades. The V clade had 53, and the GH and GR clades had 89 clades each. In every case, the RBD region was more highly conserved relative to the spike, and was lowest for clades S and V. The virus may conserve the RBD since it is essential for targeting cell infection.
Among them, the most important was Hap-1, found to be present in 54% of strains in clades G and GH, and at 56% in GR. However, another, called Hap-252, was the predominant haplotype in clades V, L, S and O, at frequencies of 70%, 63%, 52% and 40%, respectively. The lowest number of haplotypes was in clade V, as was the nucleotide diversity.
Some changes are linked to functional attributes, such as one in clade L, and the D936Y in clade GH, which are associated with monomer stability. Another, A829T, in clade S, is linked with the fusion peptide. Such changes are found in 1% to 3.4% of the sequences.
The rate of evolution of the spike protein was estimated to be 1.08 x 10-3 nucleotide substitutions/site/year, and did not change between clades. However, the rates were somewhat lower for the founding clades relative to the more recent clades G, GH and GR. The overall rate of S nucleotide evolution is higher than that found from analyses of the whole genome.
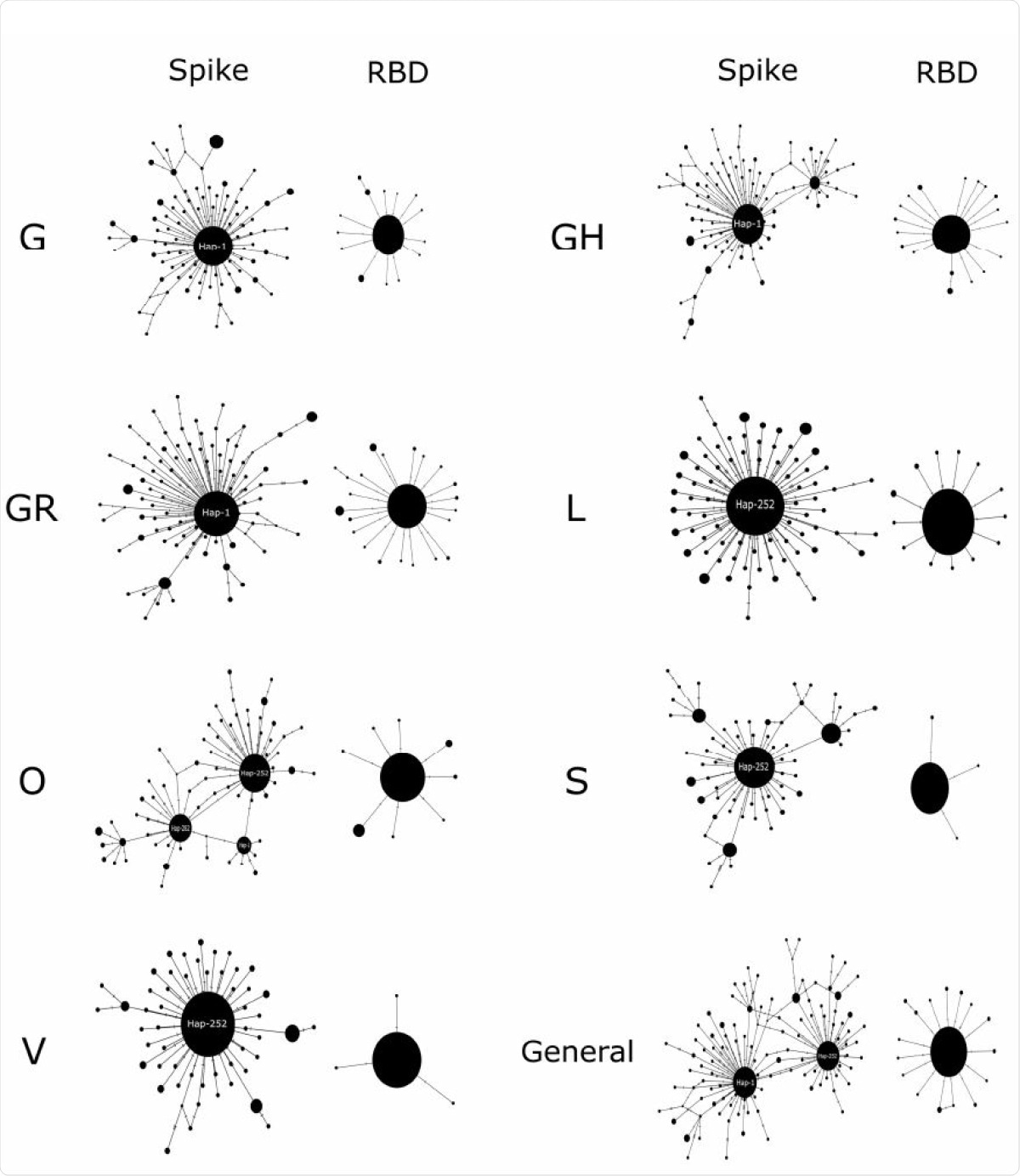
Median-joining haplotype networks. The seven clades of SARS-CoV 2 described to date are compared to both the entire Spike and the RBD coding region. The diameters of the spheres are proportional to the frequency of haplotypes. The main haplogroups are indicated.
The primary reason for this higher evolutionary rate is because the genome contains many regions that are highly conserved, but the S region, on the other hand, is among the sequences that are changing most rapidly. However, there is no marked difference between this and the result of the analysis performed over the first four months of the pandemic.
The study reports the process by which the spike protein underwent changes in different clades, while the virus was circulating in a global naïve population without pre-existing immunity. The results showed that the spike region remained quite stable over this period. As vaccines begin to be deployed, and a significant proportion of the world becomes infected and recovers, this situation could change, and require a re-evaluation of this parameter.
The authors point out, “The present evolutionary analysis is relevant since the spike protein of SARS-CoV-2 is the target for most therapeutic candidates; besides, changes in this protein could have consequences on viral transmission, response to antivirals and efficacy of vaccines.”
It could also be important in terms of understanding the characteristics of reinfection by other clades of the virus, as has already been reported.