The clinical presentation of coronavirus disease 2019 (COVID-19) ranges from mild flu-like symptoms in many patients to severe respiratory failure and death in patients with comorbidities. Although studies have shown correlations between hyperactivation of the immune system and the formation of a cytokine storm in patients with severe COVID-19, the host genetic factors that rule these immune events and determine the susceptibility of some patients to severe disease is not well understood.
.jpg)
A cytokine storm cell signaling vector horizontal background. Image Credit: WhiteDragon / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
In order to address this gap in knowledge, genome-wide association studies (GWAS) have been performed, and they have identified several genetic variants linked to COVID-19 susceptibility and severity. However, the genes they target and the immune cell types most affected by these variants are not fully clear. Identifying common genetic polymorphisms linked to the severity of COVID-19 disease can help uncover molecular pathways and immune cell types that drive COVID-19 pathogenesis.
The DICE database of immune cell gene expression, epigenomics, and expression quantitative trait loci (eQTLs) was created to answer such questions and to help specify the functional variants in dense haploblocks associated with disease susceptibility.
Assessing the effects of COVID-19-risk variants on gene expression in several immune cell types
A team of researchers from the University of California San Diego, USA, and University of Liverpool, UK, used the DICE database and 3D cis-interactome maps to generate a list of target genes and immune cell types that are most affected by the genetic variants associated with COVID-19 severity. Their study is published on the preprint server, bioRxiv*.
The researchers systematically assessed the effects of 679 COVID-19-risk variants (as defined by the COVID-19 Host Genetics Initiative) on gene expression in 13 immune cell types and two activation conditions. The expression of 11 protein-coding genes and one non-coding RNA or eGenes was linked to the genetic variants associated with severe COVID-19 requiring hospitalization. Interestingly, most of the eGenes linked to severe illness showed noticeable effects in specific immune cell types.
On applying a more liberal GWAS association P value threshold of 1 x 10-5, the researchers identified 41 additional eGenes linked to genetic variants non-significantly associated with severe COVID-19. Some of these variants might reach statistical significance as more donors with severe illness are included in the subsequent phases of the analysis.
Results show links between genetic variants and gene expression in specific immune cell types
The results of the study show that severe COVID-19-risk variants were significantly linked to the expression of 11 protein-coding genes and overlapped with cis-regulatory or target gene promoter regions that interact with target promoters in the immune cell types where their effects are most noticeable.
For instance, the researchers identified that the links between variants in the 3p21.31 risk locus and CCR2 expression in classical monocytes are likely mediated via an active cis-regulatory region that interacted with CCR2 promoter in monocytes.
Thus, our findings point to a potentially important role for IL-10 signaling and NK cells in influencing susceptibility to severe COVID-19 illness.”
The expression of many other genes suggested the presence of distinct genotype-dependent effects in non-classical monocytes, B cells, NK cells, or specific T cell subtypes. This highlights the possibility of COVID-19 risk variants impacting the function of various immune cell types and influencing severe COVID-19 manifestations.
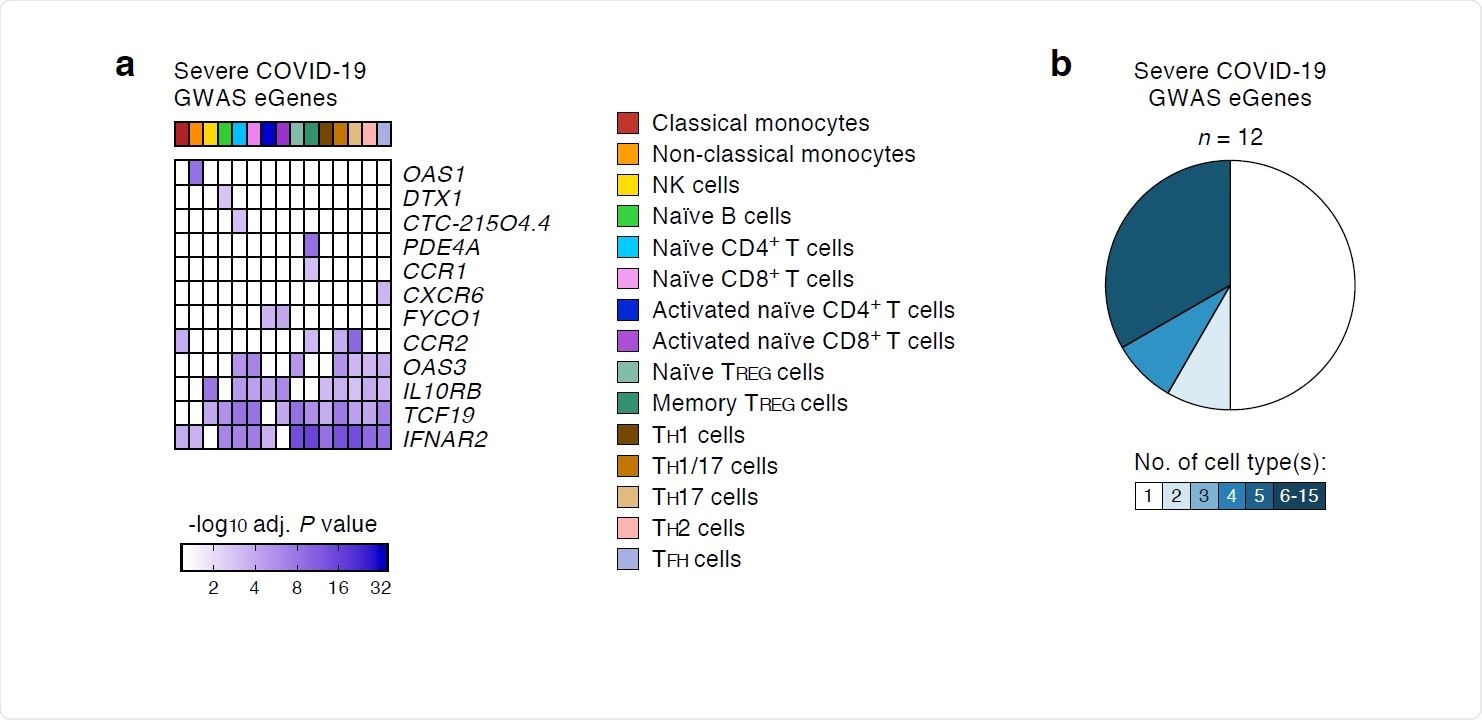
COVID-19-risk variants with eQTL activity in human immune cell types. (a) Genes and cell types influenced by GWAS SNPs linked to severe COVID-19 illness requiring hospitalization. For each cell type (columns), the adj. association P value for the peak GWAS cis-eQTL associated with the indicated eGenes (rows) is shown. (b) Fractions of GWAS eGenes linked to severe COVID-19 illness identified in varying numbers of cell types.
Genetic polymorphism data can help define pathways and cell types that drive COVID-19 pathogenesis
To summarize, many genetic risk variants of severe COVID-19 show immune cell-type-restriction of their impact on gene expression and thus may have the potential to affect the function of various immune cell types and genetic pathways.
According to the authors, their analysis of eQTLs and cis-interaction maps in diverse immune cell types enabled a definition of the immune cell types and genes that drive susceptibility to severe COVID-19 disease, which possibly contributes to the difference in clinical outcomes.
This study also highlights how common genetic polymorphism data can help define molecular pathways and immune cell types that have a role in COVID-19 pathogenesis.
Our study also highlights how information about common genetic polymorphisms can be used to define molecular pathways and cell types that play a role in disease pathogenesis.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Benjamin J. Schmiedel, Vivek Chandra, Job Rocha, Cristian Gonzalez-Colin, Sourya Bhattacharyya, Ariel Madrigal, Christian H. Ottensmeier, Ferhat Ay, Pandurangan Vijayanand (2020) COVID-19 genetic risk variants are associated with expression of multiple genes in diverse immune cell types. BioRxiv preprint server. doi: https://doi.org/10.1101/2020.12.01.407429, https://www.biorxiv.org/content/10.1101/2020.12.01.407429v1
- Peer reviewed and published scientific report.
Schmiedel, Benjamin J., Job Rocha, Cristian Gonzalez-Colin, Sourya Bhattacharyya, Ariel Madrigal, Christian H. Ottensmeier, Ferhat Ay, Vivek Chandra, and Pandurangan Vijayanand. 2021. “COVID-19 Genetic Risk Variants Are Associated with Expression of Multiple Genes in Diverse Immune Cell Types.” Nature Communications 12 (1): 6760. https://doi.org/10.1038/s41467-021-26888-3. https://www.nature.com/articles/s41467-021-26888-3.