Remdesivir, a drug used to treat patients with coronavirus disease (COVID-19), has been shown to target both RNA-dependent RNA polymerase (RdRp) and main protease (Mpro) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – resulting in a ‘double whammy’ effect against this pathogen. These findings were recently published in the Journal of Physical Chemistry.
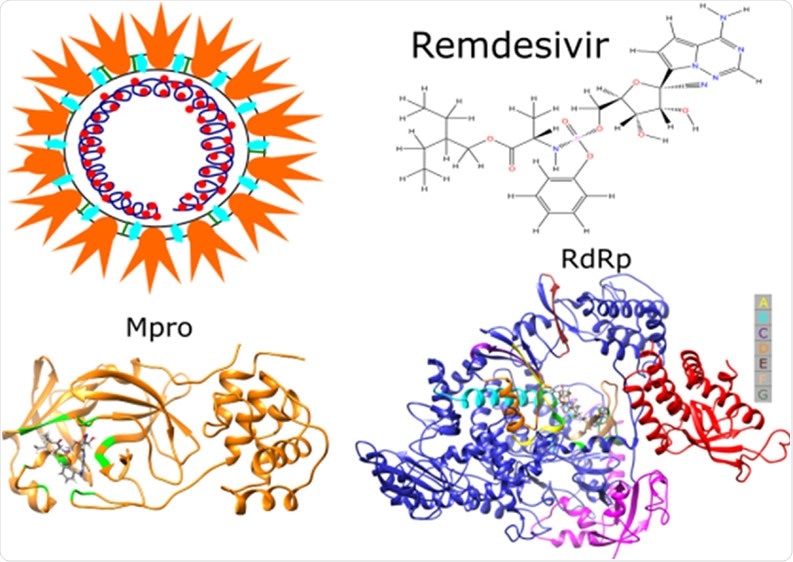
Image Credit: https://pubs.acs.org/doi/10.1021/acs.jpcb.0c07312
The escalating outbreak of SARS-CoV-2 has caused a COVID-19 pandemic, resulting in more than 66 million cases and 1.5 million deaths around the world (as of December 6, 2020). Although several promising vaccines are now entering the stage, there is still no efficacious drug at our disposal.
However, a drug repurposing quest among commercially available drugs was underway since the early days of the pandemic. Due to its initial positive results in the clinical milieu, remdesivir has been pervasively used in many countries to treat patients with COVID-19, but without full information on the exact molecular targets.
It has been shown previously that the drug can inhibit SARS-CoV and MERS-CoV viruses by interfering with RdRp. Can it also do the same when confronted with SARS-CoV-2, and what are its molecular mechanisms of interaction and possible targets?
Hence, the researchers from the Institute for Computational Science and Technology in Ho Chi Minh City in Vietnam decided to perform molecular docking, steered molecular dynamics simulations, and umbrella sampling to answer this pertinent question.
A range of computational tools
By combining the aforementioned computational tools and techniques, this research group (led by Dr. Hoang Linh Nguyen) examined the binding affinity of remdesivir to two targets, which includes Mpro (also known as 3C-like protease) and RdRp of the SARS-CoV-2.
The structure of remdesivir has been retrieved from PubChem data bank, while Mpro and RdRp structures were derived from the Protein Databank. All residues that have been missing were successfully added with the use of the MODELLER program package.
Docking simulation has been used to dock remdesivir to the Mpro and RdRp binding sites, while molecular dynamics simulation was conducted to capture motion properties and schematics of binding processes.
Finally, steered molecular dynamics proved to be valuable for obtaining relative biding affinities in this study, while umbrella sampling has been used to estimate the binding free energy of Mpro and RdRp.
Strong binding to two targets
The evaluation of binding affinities in this study has revealed that remdesivir firmly binds to both targets. Detailed results show that the drug binds to Mpro marginally weaker in comparison to RdRp. The corresponding inhibition constants fall within the micromolar range.
However, the binding mechanisms of remdesivir to these two targets are different. More specifically, the main force for stabilization of the RdRp–remdesivir complex is the electrostatic interaction, while the van der Waals interaction is dominant in the Mpro–remdesivir case.
Moreover, the per-atom distribution of the non-bonded interaction energy of remdesivir with RdRp and Mpro is similar for both complexes, and the study has shown that 10-50 atoms exhibit strong interactions.
Implications for treatment
"Our result indicates that remdesivir can target not only RdRp but also Mpro, which can be invoked to explain why this drug is effective in treating COVID-19", explain the study authors their main findings.
We have identified residues of the target protein that make the most important contribution to binding affinity, and this information is useful for drug development for this disease"
This work now awaits verification with various in vitro and (possibly) in vivo experiments. Also, the extension of this work to other targets will be pivotal not only in the domain of fundamental research but also in the realm of clinical applications.
Journal reference:
- Nguyen, H.L., et al. (2020). Remdesivir Strongly Binds to Both RNA-Dependent RNA Polymerase and Main Protease of SARS-CoV-2: Evidence from Molecular Simulations. Journal of Physical Chemistry. https://doi.org/10.1021/acs.jpcb.0c07312.