The COVID-19 pandemic is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the seventh known pathogenic coronavirus among humans. While three of these are epidemic, the rest cause endemic disease, mostly mild upper respiratory infections like the common cold. All are zoonoses.
Currently, several scientists have suggested that a significant proportion of the population have pre-existing immunity because of T cells primed against SARS-CoV-2 antigens before exposure. This is thought to be the result of prior infection by the cross-reactive endemic human coronaviruses. However, a new study published in the preprint server bioRxiv* in 2020 concludes that this may not be the case.
The surprisingly wide range of clinical severity of COVID-19 has roused much speculation about the reasons. Among the more common suggestions are the effects of comorbidities, age, sex, climatic conditions, and vitamin D levels.
Several earlier studies have looked into the presence of T cells or antibodies to SARS-CoV-2 in samples collected before the pandemic, from healthy individuals. The researchers concluded that some degree of T cell immunity could be observed in different parts of the world, in people who had never been exposed to the virus.
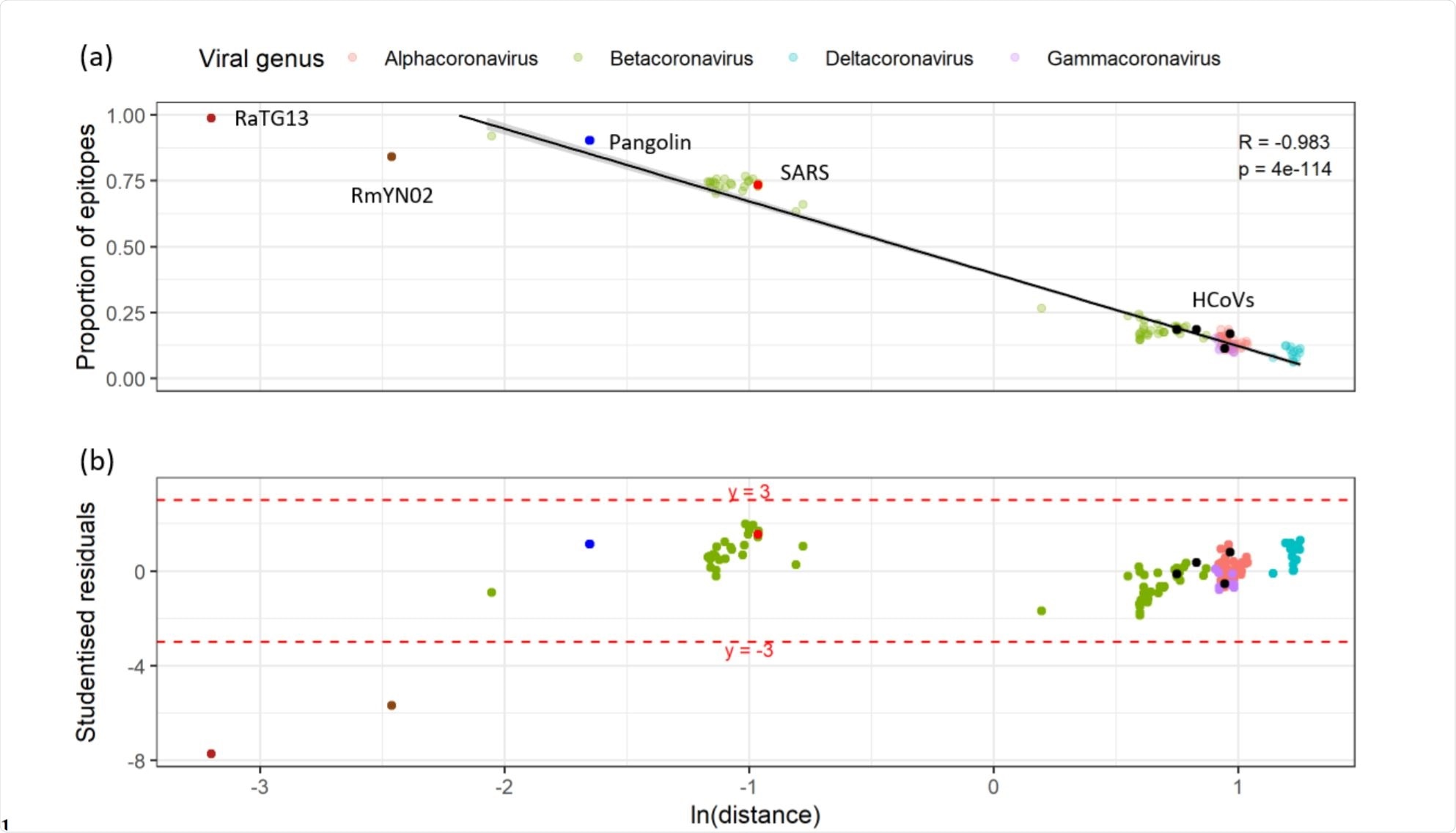
Relationship between the proportion of unexposed epitopes that have detectable sequence homology and the cophenetic distance to SARS-CoV-2 in a representative subset of the Coronaviridae. Image Credit: https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1.full.pdf

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Study Details
The current study attempted to look at the contribution of the seasonal coronaviruses, if present, and how this affects the disease progression. For instance, some think that children, who are known to frequently suffer from the common cold, are thereby protected against severe disease. Others go so far as to suggest that there is another zoonotic coronavirus circulating at present, which, though unknown, is also modifying the expected spread and pathogenicity of SARS-CoV-2.
The researchers relied on bioinformatics for their study, using samples from healthy individuals obtained before the onset of the outbreak. They constructed a core- or shared-gene-based phylogenetic tree covering all known sequenced coronaviruses. The four core genes used were ORF1ab, S, M, and N. the data came from over 2,500 coronaviruses. This was used to measure how far the 177 cross-reactive T cell epitopes (antibody binding sites) identified in such individuals could be said to correspond to those of endemic human coronaviruses and other coronaviruses, in general.
At thresholds of sequence identity set at 40%, 60%, and 80%, the peptide sequences of SARS-CoV-2 are found to be highly conserved across the whole coronavirus family. Homologous regions include the RNA-dependent RNA 101 polymerase (RdRp) (nsp12) and helicase (nsp13).
Low Homology with Endemic Coronaviruses
Surprisingly, less than half the T cell epitopes passed this test concerning endemic viruses. From 76% to 83% of SARS-CoV-2 epitopes had no obvious homology with any of the four latter, when compared individually, while almost 60% remained non-homologous when their epitope sequences were compared with those of all four in combination. These were termed ‘unexplained epitopes’, as a result.
When compared to the remaining sequenced coronaviruses, many of these unexplained epitopes appear homologous to some beta coronaviruses, including the earlier SARS-CoV.
Observed Homology Corresponds to Genomic Relationship
The researchers then prepared a smaller subset of 155 T cell epitopes, containing one host and one viral representative from each species. This analysis showed that the number of published epitopes with observable homologous sequences to any of the other sequenced coronaviruses is proportional to the natural logarithm of the phylogenetic distance of that virus from SARS-CoV-2. In other words, very closely related viruses showed the highest degree of conservation of sequences.
On the other hand, the number of reported epitopes with significant homology to any of the coronavirus sequences available at present was not greater than could be attributed to chance alone. Moreover, the highly-homologous sequences, covering nsps12-16, were not primary T cell targets in patients who had recovered from COVID-19.
Again, SARS-CoV-2 typically causes a multifaceted immune response covering the whole genome, mostly outside the spike protein, not focused on the terminal region of the highly conserved ORF1ab, and is not associated with cross-reactivity with endemic human coronaviruses.
No Single Candidate
The researchers then compared the 177 epitopes with the NCBI database of proteins from all kinds of species. Taking only the first 1,000 hits in each category, they found that 10 of the epitopes share their sequences partially with proteins from a broad range of organisms, belonging to very different classes such as viruses, bacteria, and eukaryotes. On the other hand, this is not higher than that due to sheer chance.
Together with the wide diversity of taxa, 170 identified, the results suggest that there is no single candidate for the source(s) of the T-cell cross-reactive repertoire beyond the Coronaviridae.”
What are the Implications?
The researchers conclude that rather than T cell cross-reactivity being the effect of prior infection by seasonal or other coronaviruses, the real cause is likely to be a lifetime of exposure to a large and variable range of microbial antigens. Nor is it likely to be the result of a widely circulating but undiscovered coronavirus which has now disappeared, and which was not genetically related to any known coronavirus.
The most plausible candidates for the cross-reactive T cell epitopes are major vaccines or extensively distributed microbes. The same picture in HIV and influenza has been attributed earlier to prior exposure to environmental antigens.
The existence of prior immunity to a newly emerging agent may thus be due to immunity elicited by other agents. In the current context, further research is required to estimate the contribution of such cross-reactivity to immunity against SARS-CoV-2, with respect to protection against infection and severe disease.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources