The second wave of the coronavirus disease 2019 (COVID-19) has hit many countries hard, with infection rates and mortality comparable to that of the first wave, or even exceeding them, in some cases. Despite the roll-out of the first vaccines, there is a need for effective antivirals to treat patients with severe disease, as well as to forestall a similar situation in future pathogenic coronavirus pandemics.
Structural basis of SARS-CoV-2 RdRp complex stalling by remdesivir. Image Credit: https://www.biorxiv.org/content/10.1101/2020.12.14.422718v1.full.pdf

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Remdesivir and viral inhibition
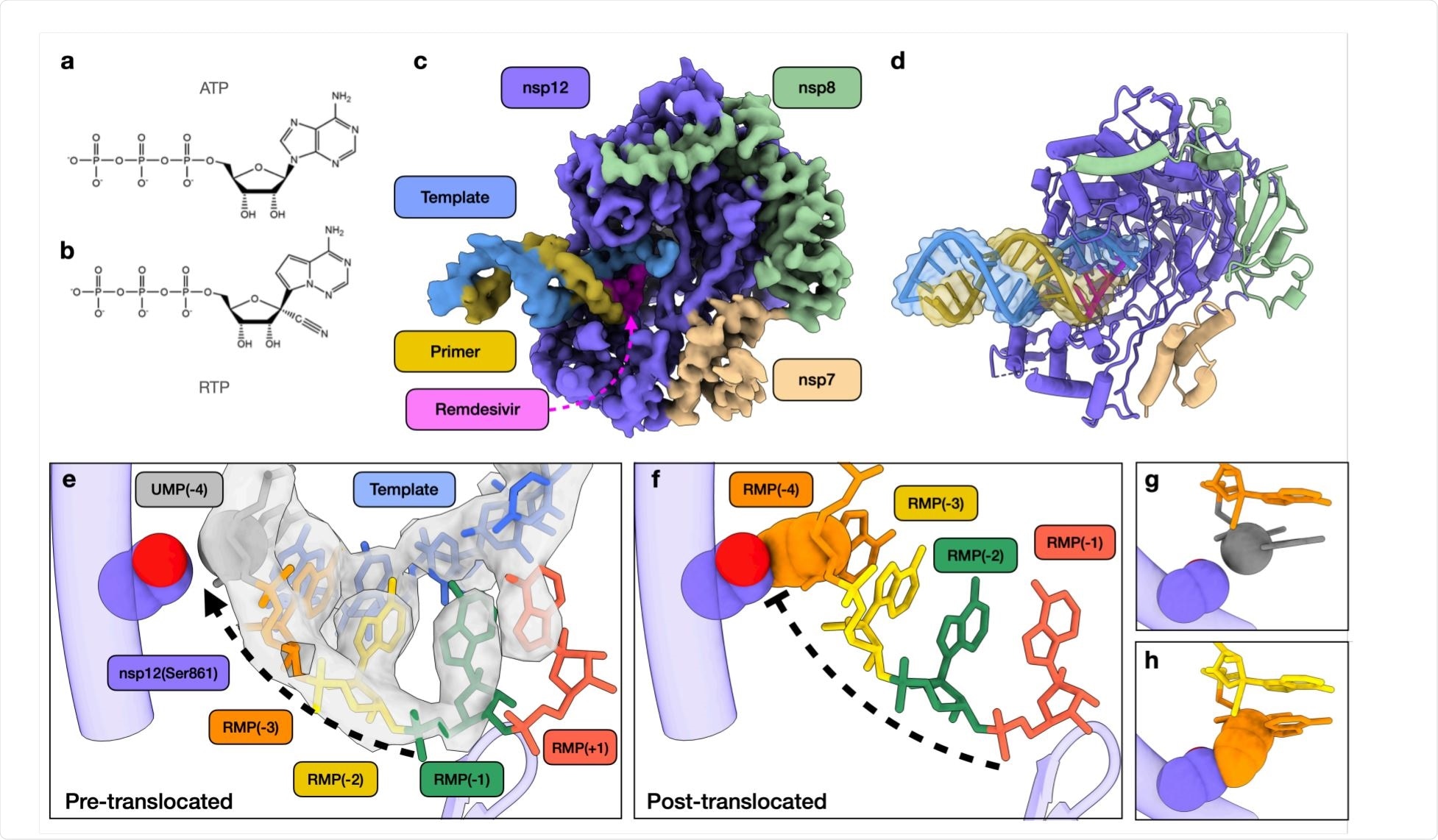
This virus has an envelope studded with the trimeric spike glycoprotein that forms the ‘corona’ of its name, and a large RNA genome. Upon viral entry into the host cell, it forms an RNA-dependent RNA polymerase (RdRp) complex consisting of the non-structural protein nsp12, and the accessory proteins nsp7 and nsp8 proteins.
One of the few approved drugs to treat COVID-19 is remdesivir, an adenosine analog. This drug appears to block the activity of RNA-dependent RNA polymerase (RdRp). The structure of this complex has been visualized to understand how RNA replication proceeds. However, the mechanism of action of remdesivir remains unclear.
Study details
The current study describes the structure of the RNA-dependent RNA polymerase (RdRp) complex after the incorporation of remdesivir, revealing how this drug acts.
Firstly, the researchers incubated the RdRp with a primer-template RNA duplex to allow it to incorporate two uracil triphosphate and three remdesivir triphosphates (UTP and RTP, respectively) molecules at high speed, within one second. This has already been demonstrated in earlier research.
On continuing the observation, they found that a fourth RTP was incorporated at a much slower rate, at 0.13/second. No further incorporation took place, which suggested the complex was stalled.
They found that the mixture consisted of 10% and 90% of 25-nucleotide and 26-nucleotide products, incorporating 3 and 4 remdesivir monophosphates (RMPs), respectively. In these conditions, when UTP and ATP are added to the solution, the result was a 27-nucleotide product, showing that premature stalling did not occur. When all four nucleotides were added to this product at 250 µM, the product rapidly grew to the full length of 40 nucleotides in 15 seconds.
Conversely, when added to the stalled RNA, an extension to 40 nucleotides occurred in only 70% of the 26-nucleotide primer, at a slow rate of 0.08/second, taking 15 seconds. This is 4,000 times slower than the normal rate of incorporation. These results agree with earlier studies. However, with the current very active replicating enzyme, a further observation arises, namely, that the inhibition can be overcome by incubating the product with high nucleotide concentrations.
When the intermediate products were examined under cryo-electron microscopy, the researchers were able to model a primer elongation intermediate stalled by remdesivir. At the active site of the RdRp complex, they found that RMPs were present at 1, 2, and 3 nucleotides downstream of the active site. However, there was an additional density in the polymerase active site, that indicates the presence of another remdesivir.
The fourth RMP stalls translocation
This confirms the earlier analysis and suggests the formation of base-pairing between the fourth RMP and the uracil monophosphate (UMP) of the template as well as stacking interactions with adjacent RMPs.
However, this may create a certain amount of disorder in the RNA backbone. It was unexpected to find the fourth RMP at the active site of the enzyme rather than translocating to allow the binding of the next UMP. This step, therefore, seems to be responsible for the stalling of the enzyme.
The researchers created a model of a putative stalled RdRp, with the first RMP moving down to the fourth position downstream of the active site. In this case, this RMP would be sterically hindered by the side chain of serine at position 861 of nsp12. Thus, this serine physically prevents or slows the translocation of the primer: template complex, and may also disrupt the normal structure of the active site. As a result, the rate of chain elongation slows down.
This model, therefore, explains the structural changes that lead to RdRp stalling following the addition of the fourth RMP. Earlier studies have shown that mutations of Ser861 on nsp12 reduce the activity of remdesivir, agreeing with the current findings.
Again, this study demonstrates the ability of the four cyano groups of the four RMPs to fit within the active site. Steric clashes occur only when the RNA duplex tries to shift at the fourth RMP, to free the active site to bind and add another UTP, supporting similar findings from another recent study.
What are the implications?
This study is the first to reveal the structure of a stalled elongation complex of an active RdRp complex.
The structural data together with the kinetic data define the mechanism of inhibition by remdesivir as a delayed chain extension inhibitor.”
The researchers hypothesize that in the presence of the proofreading exonuclease, nsp10/14, the 3’ terminal base would be removed from the stalled enzyme and rapidly replaced. This could end in a steady state with repeated removal and replacement of this base. As a result, further elongation of the RNA would be inhibited while preventing the excision of the RMP.
In support of this, mutations that reduce exonuclease activity enhance the effect of remdesivir fivefold. Possibly, if the exonuclease is inactive, remdesivir may become buried in the viral genome and cause lethal mutations to arise.
Our current data provide a structural basis for RMP-induced stalling, which may guide the development of more effective nucleoside analogs.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources