The extent of the coronavirus disease 2019 (COVID-19) pandemic is staggering. Among the worst aspects of the disease is the immense uncertainty about the clinical course, with some developing severe or critical COVID-19, and often dying of it, while others remain asymptomatic or have only mild symptoms throughout. A new preprint appearing on the medRxiv* server in December 2020 describes some potentially important genetic factors in the host that may shape the clinical symptoms and signs of the disease.
A recent study reported the outcome of a genome-wide association study (GWAS) that showed two genetic loci possibly linked to increased susceptibility to the disease process. The first was on chromosome 3, encoding a number of genes that may have a functional role in COVID-19 disease. The second was on chromosome 9, which also harbors the ABO blood group. A second recent study demonstrated the presence of uncommon gene mutations in the type I interferon pathway that contribute to the risk of serious or critical pneumonia in COVID-19. Even more recently, two separate GWAS reports showed evidence that the first two susceptibility loci were actually associated with high risk for serious disease. These studies also identified new variants on severe loci on chromosomes 19, 12 and 21 that were linked to severe COVID-19.
Study details
The current study aimed to explore the possibility that there are some variants on certain loci on chromosome 21 near the genes that encode TMPRSS2 and MX1. They examined GWAS data from the COVID-19 Host Genetics Initiative, and mapped chromosome 21 in deep mapping. They found that, indeed, commonly found variants were significantly associated with severe COVID-19.
In a large cohort of over 908,494 individuals, from Europe, the researchers found five single nucleotide polymorphisms (SNPs) at the TMPRSS2/MX1 locus, which were associated with less severe disease. The cohort included ~6,400 severe cases and over 9,00,000 controls. The same association was found in two Asian cohorts, with all five SNPs, and two and one SNPs were confirmed in African and Italian cohorts, respectively.
Using the expression quantitative loci (eQTL) parameter to assess the relationship between the gene variants and gene expression, of the most significantly disease-associated SNPs, they found that all the top five SNPs showed eQTL signals for MX1 in the blood. The minor alleles of these variants, the gene less often expressed, was correlated with higher MX1 levels. MX1 is a guanosine triphosphate (GTP)-metabolizing protein that takes part in cellular antiviral responses, inhibiting viral replication. It is induced by both type I and III interferon pathways.
Of the other SNPs, 9/14 did not show such signals, except for one. The minor alleles of 4/5 SNPs also correlated inversely with TMPRSS2 expression. One of these minor alleles is, in addition, an eQTL for TMPRSS2 in osteoblasts following treatment with dexamethasone.
The uncovering of host genetic factors that may explain the marked differences in COVID-19 clinical features is important, not just to understand the pathogenesis of the disease, but also to identify therapeutic targets that will respond to drugs. Human host cells express both angiotensin-converting enzyme 2 (ACE2) as well as the transmembrane serine protease TMPRSS2. The authors postulated that genetic variants with both reduced TMPRSS2 and higher MX1 may be connected with a better outcome following SARS-CoV-2 infection.
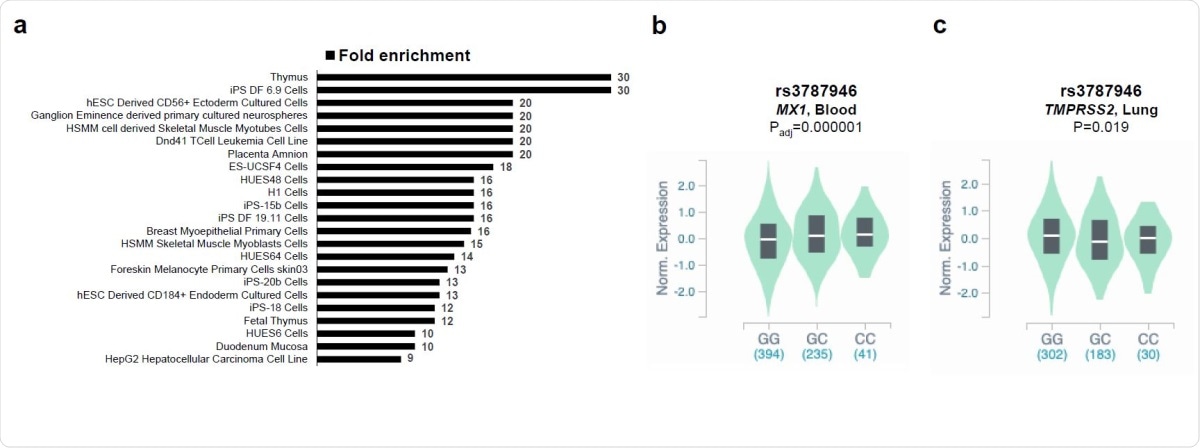
Enrichment of SNPs in regulatory regions and eQTL analyses. The statistically significant fold enrichments (P<0.05 after Bonferroni correction) of SNPs in regulatory DNA regions active in different tissues are shown (a). eQTL violin plots between genotypes of rs3787946 (b) and rs3787946 (c) with MX1 and TMPRSS2 expression from the from the Genotype-Tissue Expression (GTEx).
What are the implications?
Not only did these five SNPs show a high frequency, but minor alleles were far less common among more severe cases, suggesting they play a protective part. The reproducibility of these SNPs in multiple cohorts suggests that the q22.3 locus on chromosome 21 is “a novel susceptibility locus to unfavorable outcome of COVID-19 and suggest that molecular mechanisms underlying this genetic predisposition may be common among individuals with different ethnicity.”
The SNPs here are enriched in the regions that regulate pluripotent stem cells and thymus. The latter is connected to adaptive immunity, but undergoes functional waning with age. The inflammatory tendency associated with immune senescence of aging is a key factor in many immune responses. It is significant that both aging and inflammatory age-related diseases are associated with severe COVID-19. The association of these gene variants with a dysfunctional thymus could be another explanation for this link with severe COVID-19.
The findings also show that MX1 is possibly related to the clinical phenotype in COVID-19, supporting the conclusions of a recent study that suggests this molecule is an effector against SARS-CoV-2. Other supportive findings include a higher MX1 expression in infected individuals and in those with higher viral loads. The MX1 levels are lower with advancing age.
Mutations that inactivate the type I interferon pathway are recurrent in COVID-19 patients. In this light, the current study supports “the evidence that the use of drugs, activating IFN signaling, could be an effective treatment to prevent the adverse outcome of disease.” While type I or III interferons are being used in clinical trials to prevent or treat COVID-19, they may enhance or induce a cytokine storm with deadly results. Thus, “it is highly relevant to spawn new drugs that have the capacity to boost the host immune response while controlling tissue damage.”
The rs12329760 coding variant is found to be linked to lower TMPRSS2 in osteoblasts following dexamethasone treatment. This drug is a powerful anti-inflammatory. This variant is uncommon among Chinese COVID-19 patients with critical illness. Modeling predicts that it may reduce viral entry into the cells by destabilizing TMPRSS2 and ACE2 binding. Thus, these SNPs may impact the functions of TMPRSS2 as well. Uncovering the functions of these MX1-regulatory gene variants may be of great help in initiating new therapeutic avenues for this lethal condition.