By conducting a state-of-the-art interactome study between the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and host cells, researchers from the University of Texas MD Anderson Cancer Center in Houston identified 437 human proteins as the high-confidence interacting proteins – with substantial implications for understanding coronavirus disease 2019 (COVID-19) pathology and potential treatments. The study is currently available on bioRxiv* preprint server.
Considering the transmission, morbidity and mortality patterns of the COVID-19 pandemic, it is evident that SARS-CoV-2 is a pathogenic coronavirus that spreads easily through the air. And as the vaccination kicks off in many countries of the world, the quest for viable treatment continues.
One of the key facets of SARS-CoV-2 lifecycle is host-virus protein-protein interactions. As these may unveil promising drug targets, the analysis of host-virus interactome (i.e., a complete set of molecular interactions in a particular cell) is direly needed and have actually been reported recently.
For that purpose, we have two well-established strategies for studying protein-protein interactome on our disposal – affinity purification and a proximity labeling-based strategy, which is followed by mass spectrometry analysis. Consequently, it was recently shown that the combination of these methods is the optimal way to generate comprehensive insights.
With this in mind, a research group led by Dr. Zhen Chen from the Department of Experimental Radiation Oncology at the University of Texas MD Anderson Cancer Center in Houston (USA) started with a quest for key human proteins that are implicated in the SARS-CoV-2 life cycle.
Two methods for an exhaustive interactome
In this study, the researchers applied the two aforementioned strategies: tandem affinity purification with the SFB (S-protein, FLAG epitope, and streptavidin-binding peptide) tag, as well as proximity labeling by using a second-generation biotin ligase, BioID2.
Genome annotation disclosed 29 SARS-CoV-2 gene products – including 16 non-structural proteins, 4 structural proteins and 9 accessory factors. Furthermore, stable viral gene expression was done in cells and subsequently verified by immunoblotting technique.
The identified proteins were filtered with the use of Significance Analysis of INTeractome (abbreviated as SAINTexpress). The researchers have also built an interaction network by utilizing the 437 identified virus-host protein-protein interactions, which enabled all the complex analyses that they have pursued.
High-confidence interacting proteins
In short, from a total of 437 high-confidence interacting proteins that bind to one or more SARS-CoV-2 genes, the researchers have identified several gene products, M protein, NSP6, ORF3a, ORF6 and ORF7b that interacted with host cell membrane proteins and complexes.
The transmembrane domain prediction also indicated that these viral gene products contain at least one transmembrane domain in their protein sequences – with the exception of ORF6, which is actually a short protein with only 61 amino acids.
Moreover, 314 high-confidence interacting proteins were identified from tandem affinity purification experiments, while 130 of them with the use of BioID2 strategy. Interestingly, only seven proteins overlapped between these two methods.
When the interactomes of NSP1 and N protein (i.e., two key SARS-CoV-2 proteins) were compared with other human coronaviruses, host pathways manipulations and divergent protein-protein interactions responsible for differences in disease pathology were uncovered.
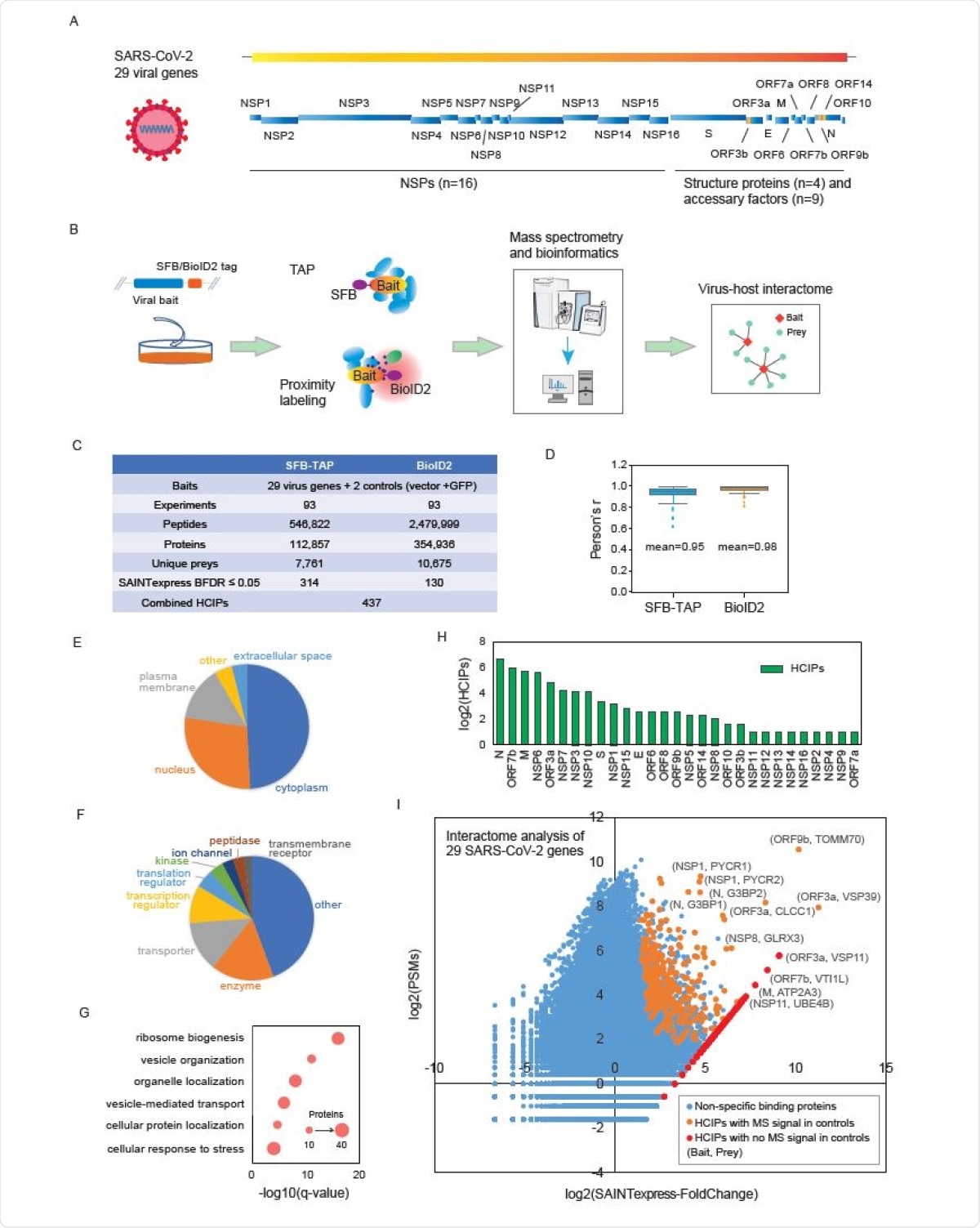
Summary of the SFB-TAP and BioID2 interactome experiments. (A) SARS-CoV-2 genome annotation, predicting 29 virus gene products. The 16 non-structure proteins (NSPs) are cleaved products of the large polyprotein open reading frame (ORF)1ab or ORF1a. These polyproteins are cleaved into small function fragments or NSPs after translation. (B) Workflow for the comprehensive virus-host interactome analysis. Two different labeling strategies, SFBTAP and BioID2 labeling, were applied in the study. Samples were analyzed by Q Exactive HF mass spectrometry (MS). (C) Summary of the datasets obtained from SFB-TAP and BioID2 results, including the number of high-confidence interacting proteins (HCIPs). BDFR, Bayesian false discovery rate. (D) Pearson correlation coefficient among three independent biological replicates of the SFB-TAP results and the BioID2 labeling experiments. (E-G) GO analysis. GO enrichment was performed using Ingenuity Pathway Analysis. Protein localization (E), molecular function (F), and biological function (G) are plotted in a single panel. (H) HCIPs identified in the purification of each SARS-CoV-2 gene. (I) Correlation between peptide-spectrum matches (PSMs) of identified proteins and their fold change calculated by SAINTexpress.
Potential drug targets
Our systemic study of the SARS-CoV-2 protein-protein interaction network provides useful data on viral gene/protein functions and potential underlying mechanisms, which could lead to the identification of new drug targets for the treatment of COVID-19", say the authors.
The obtained interactome dataset not only proved some previously established host-virus interactions, but also uncovered manifold novel interacting proteins that may be pivotal for all relevant components of SARS-Cov-2 lifecycle.
In any case, findings from this study will undoubtedly be exploited in our ongoing fight against COVID-19; however, they may also suggest new ways to combat any potential coronavirus diseases in the future, which is continuously a realistic threat.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Chen, Z. et al. (2020). Comprehensive analysis of the host-virus interactome of SARS-CoV-2. bioRxiv. https://www.biorxiv.org/content/10.1101/2020.12.31.424961v1,https://doi.org/10.1101/2020.12.31.424961.
- Peer reviewed and published scientific report.
Chen, Zhen, Chao Wang, Xu Feng, Litong Nie, Mengfan Tang, Huimin Zhang, Yun Xiong, Samuel K Swisher, Mrinal Srivastava, and Junjie Chen. 2021. “Interactomes of SARS‐CoV‐2 and Human Coronaviruses Reveal Host Factors Potentially Affecting Pathogenesis.” The EMBO Journal 40 (17). https://doi.org/10.15252/embj.2021107776. https://www.embopress.org/doi/full/10.15252/embj.2021107776.