The ongoing coronavirus disease 2019 (COVID-19) pandemic continues to spread, with the virus responsible for it, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) undergoing many mutations that have affected its infectivity. Now, an intriguing recent study appeared on the bioRxiv * preprint server describes the use of in vitro evolution to elicit affinity maturation of the viral receptor-binding domain (RBD) of the spike protein to bind the host cell angiotensin-converting enzyme 2 (ACE2) with greater affinity.
The virus infects via airborne particles, in droplets, and on infected surfaces. It interacts with the receptor via its RBD. This results in the spike glycoprotein cleavage by the host serine protease, TMPRSS2, to generate a fusion peptide that mediates virus-cell membrane fusion. This allows the virus to enter the host cell. The use of specific antibodies to block the ACE2 receptor prevents virus entry, therefore.
The binding affinity of the spike protein for the ACE2 receptor is at ~10 nM, which is about ten times higher than the affinity of the earlier SARS-CoV virus, and this may be partly responsible for the higher infectivity of the current virus. This is supported by the sequencing of late mutations from South African and British variants, which seems to enhance binding to the receptor.
Now that several vaccines claiming high efficacy have been rolled out, many countries are beginning the process of immunizing their citizens. Nonetheless, since 100% protection is not available, and since the older and sicker part of the population is at particularly high risk for the disease, especially from newly emerging variants, the need for effective antivirals to be developed remains. Some of the potential targets for such drugs include the spike protein, the ACE2 receptor, and TMPRSS2.
One notable approach has been the development of neutralizing high-affinity antibodies, soluble human recombinant ACE2 or repurposed TMPRSS2 inhibitors. The current paper also deals with the ability of the RBD itself to inhibit the ACE2 receptor binding site competitively. This can happen only if the affinity of the RBD is much higher, at picomolar levels.
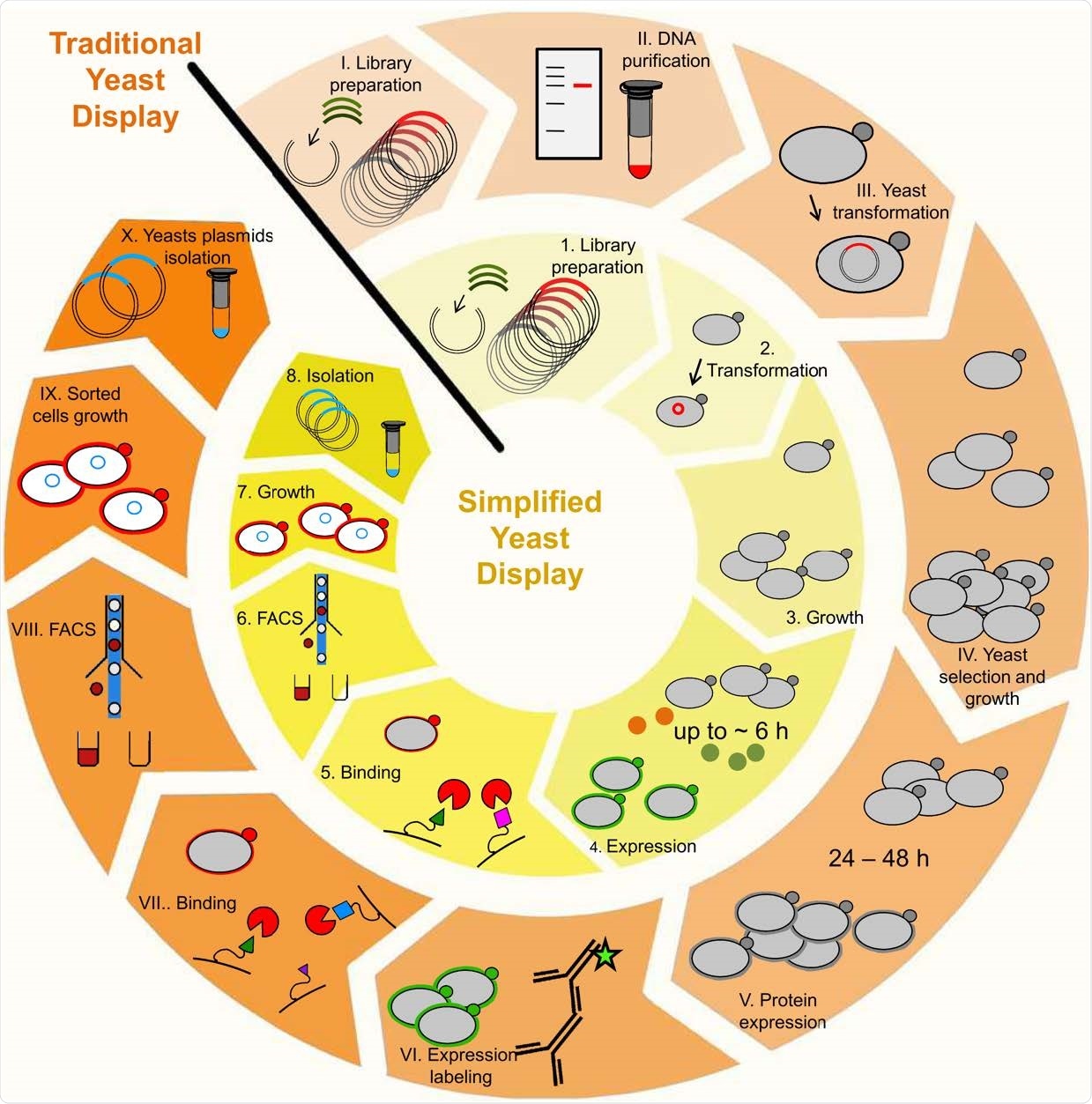
Enhanced yeast display benefits over traditional method. The use of enhanced yeast display enables elimination of DNA purification procedures between libraries (step II.); exclusion of antibody-based expression labeling procedure (step VI.), and the bright reporters eUnaG2 (orange points, step 4.) or DnbALFA (green points, step 4.) allow for ultra-tight binding selection, with reduced background and increased sensitivity in a reduced time frame (20).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Study details
The researchers used a yeast display platform to select mutations in the RBD for high-affinity binding to ACE2. This helped them to identify the most abundant among the naturally occurring RBD mutations, namely, S477N, E484K, and N501Y. These are associated with various variants like the South African (E484K, N501Y) and British (N501Y), variants that have a 13-fold and a 3.5-fold higher binding affinity for ACE2, compared to the wild-type variant. The S477N mutation is characteristic of the European mutations. The South African has the highest binding affinity, among all these, at 126 pM, relative to 1.6nM for the wild-type virus.
Mutations linked to prevalence
When they plotted the mutations in the Global Initiative for Sharing All Influenza Data (GISAID) database against the apparent change in the binding affinity of the RBD variant to the ACE2 receptor, by deep mutational scanning, they found that the greater the prevalence of the mutation, the higher the binding affinity at that position.
In short, these mutations were not randomly selected, but the most abundant variant in the population has the highest binding affinity at the mutated position. With E484R, two nucleotides in the same codon must be changed, which will require multiple random mutations to occur. This accounts for its slow spread but will not lead to its dying out.
Secondly, they also found that mutations that become abundant in the population do not impact the protein's stability, which indicates that protein stability is an essential limitation on the type of mutation that is selected.
They then examined the possibility of achieving a much higher affinity of binding. This line of investigation would help predict some future lines of SARS-Cov-2 evolution. In parallel, a very tight binder could be used to block ACE2 receptors and prevent viral binding as well.
The researchers then carried out three more rounds of yeast display to generate an enriched library of high-affinity RBDs, with several accumulated mutations. Most of these were single-nucleotide changes, but some required changes at two nucleotide positions. The latter was found at the periphery rather than at the binding interface, in agreement with earlier computational fast association (FA) library design.
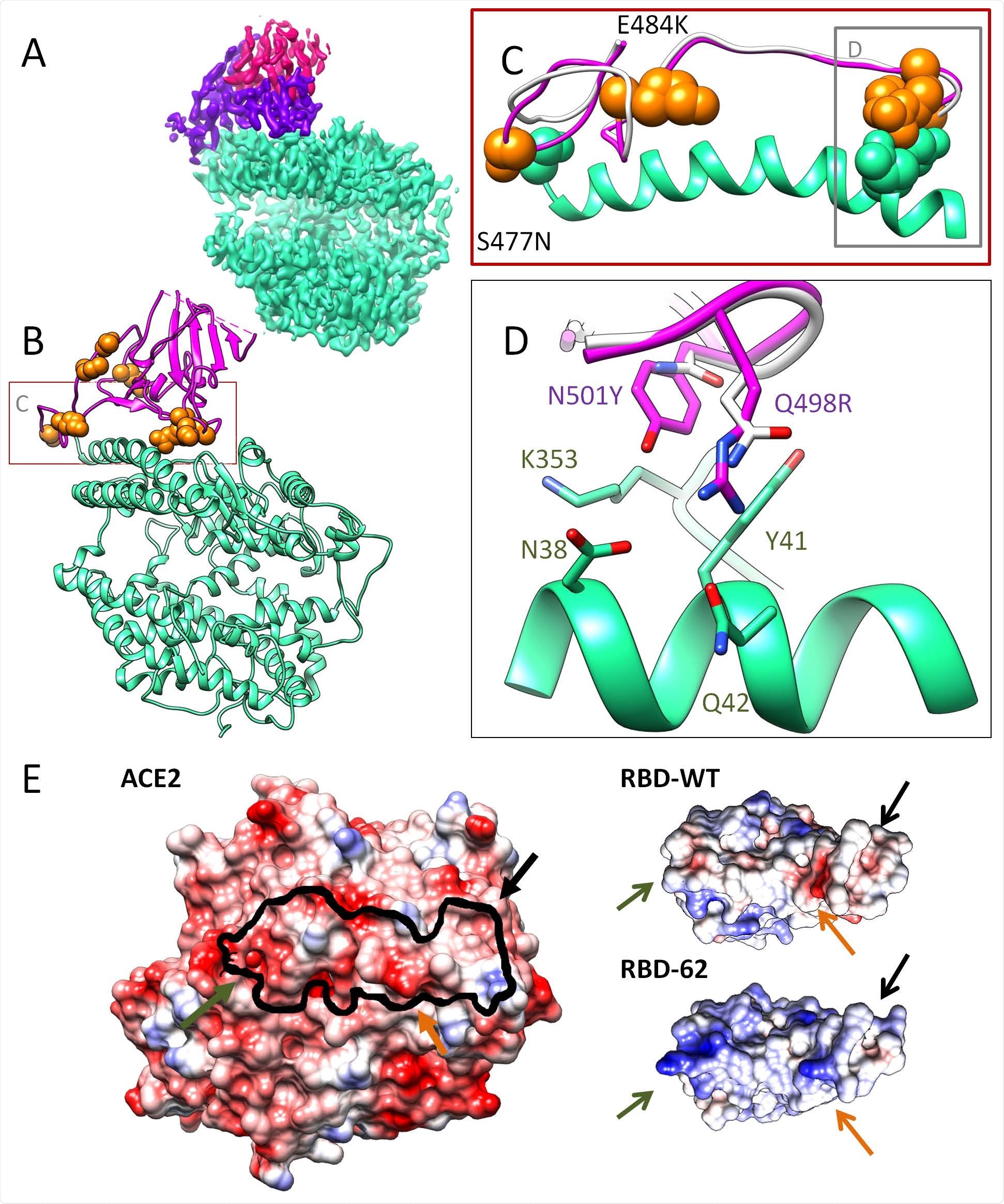
Cryo-EM structure of the ACE2-RBD-62 complex at 2.9 Å resolutions. A) The Cryo-EM electron density map with ACE2 (cyan), RBD-62 RBM (magenta), and RBD core (pink). B) Cartoon representation of the ACE2-RBD-62 model with eight mutations resolved in the electron density map (orange). C) The S477N, Q498R and N501Y mutations depicted in RBM (orange spheres) interacting with S19, Q42 and K353 of ACE2 respectively (cyan spheres) are situated at the two extremes of the RBD-ACE2 interface, suggested to stabilizing the complex. D) The interaction network formed between RBD-62 mutations and ACE2. RBD-WT residues are in white (heteroatom coloring schema). E) Electrostatic complementarity between RBD and ACE2 is strengthened in RBD-62 by positive charges at positions N460K, E484K, and Q498R. The black line one ACE2 indicates the RBD binding site.
RBD-62 blocks viral entry at low concentrations
The researchers found that one of the clones, RBD-62, had a 600-fold increase in affinity over the wild-type RBD, accompanied by a 4 0C increase in thermostability. The RBD-62-ACE2 complex was structurally similar to that of the wild-type RBD. RBD-62 has nine mutations, and three of its segments are not stabilized by its contacts with ACE2 because they are on the opposite side. The most significant differences were observed on the receptor-binding motif (RBM). S477N, Q498R, and N501Y were three of the mutations that stabilize the complex, being at the two ends of the binding interface. RBD-62 also has a much more positive surface, allowing stronger binding to the negatively charged ACE2 RBD binding surface. The N477 mutation also interacts with the S19 residue on the ACE2.
RBD-62 also does not inhibit the catalytic activity of ACE2, which is crucial in its protective role in the human body via the renin-angiotensin-aldosterone system (RAS). Finally, they found that both wild-type RBD and RBD-62 inhibited viral entry, reducing the half-maximal effective concentration (EC50) from the near-90 picomolar range to the below-10 picomolar range.
RBD-62 also prevented viral entry into and replication in over 99% of the cells, compared to only 75% with the wild-type RBD. "The complete blockage of viral replication, using a low nM concentration of RBD-62 makes it a promising drug candidate."
The use of combinatorial selection has allowed information to be gathered on the most important mutations, which can be used to identify emerging mutations. The researchers predict the possibility that the E484R variant will gain dominance, especially if it is coupled with the N501Y mutation, but the S494P mutation will not spread rapidly.
The Q498 R mutation requires a tyrosine to be added at position 501, and this paired mutation increases the affinity to below 100 pM. This position should therefore be watched carefully, for its imminent appearance, especially as R498 is in a hypervariable location of the RBD.
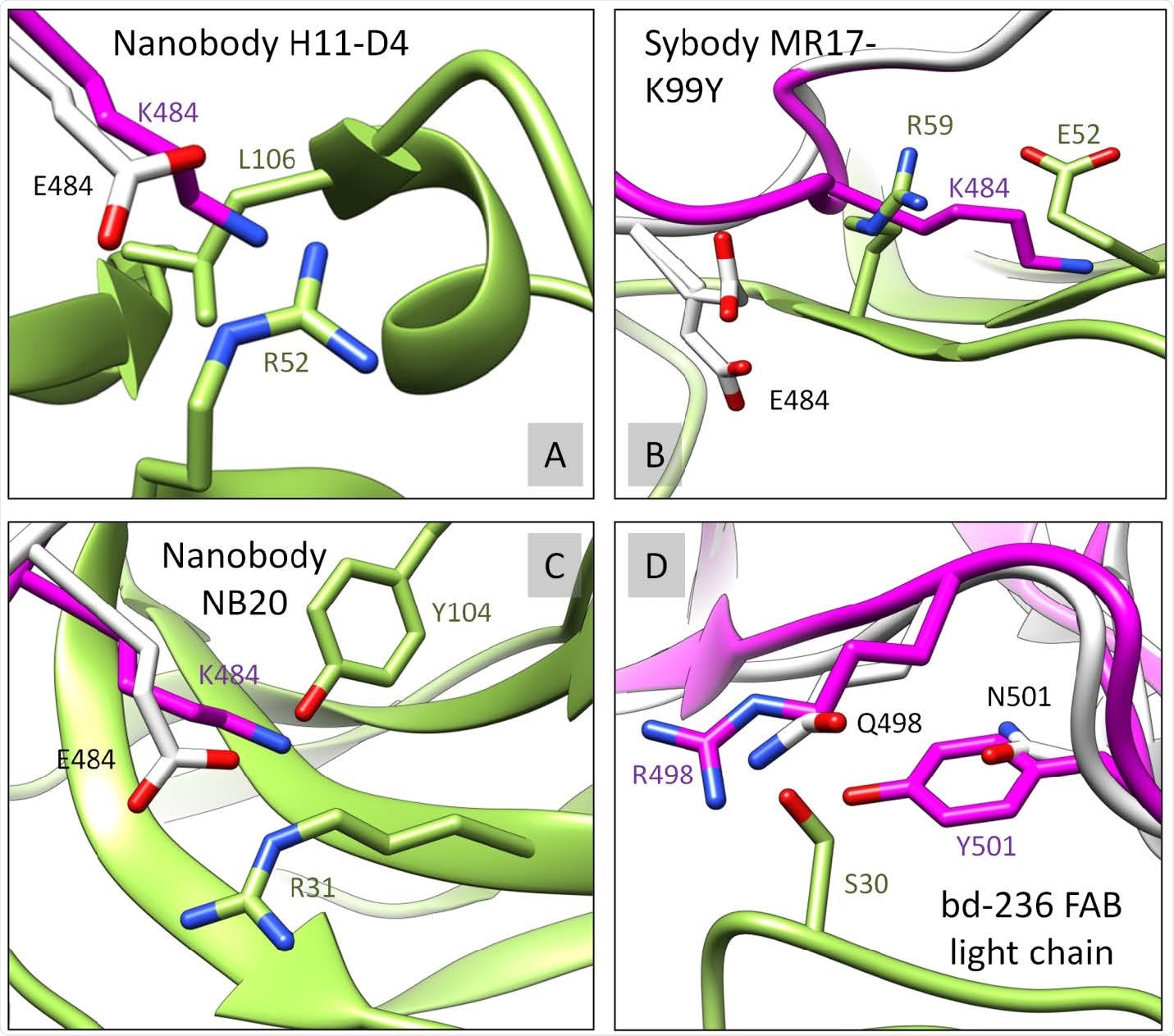
S12 RBD-62 mutations are interfering with binding to multiple antibodies. The RBD-62 (magenta) was structurally overlayed with RBD-WT (white). S477N, E484K, Q498R, and N501Y RBD mutated residues were analyzed for disruptive contacts/clashes with corresponding binding antibody/nanobody (green) in relation to RBD-WT. Four examples A) PDB ID: 6YZ5, B) PDB ID: 7CAN, C) PDB ID: 7JVB, D) PDB ID: 7CHE, where RBD-62 (but not RBD-WT) forms serious clashes with the second chain. Further experimental evaluation is needed to support our observation.
What are the implications?
RBD-62 is, therefore, "not only [a] valuable source of information but also a "crystal ball" to predict future virus evolution steps." It contains many currently circulating mutations. When tested for antibody binding, it reduces the number of contacts for 56/92 antibodies, while in nine cases, there is major steric hindrance. Most of these effects are due to E484R and Q498R. This indicates that drugs and vaccines must be closely followed up for their ability to bind to RBDs carrying current and future mutations.
And finally, the researchers advise immediate work to understand whether poorly-designed face masks are just allowing the spread to these mutations with higher binding affinity in the population. Such masks reduce the virus's concentration in exhaled air, stimulating the spread of those variants with higher binding affinity rather than the wild-type virus. If so, high-quality face masks should be made mandatory to keep exhaled viral titers too low to produce infection, as with the N95 masks.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Zahradnik, J. et al. (2021). SARS-CoV-2 RBD in vitro evolution follows contagious mutation spread, yet generates an able infection inhibitor. bioRxiv preprint. doi: https://doi.org/10.1101/2021.01.06.425392. https://www.biorxiv.org/content/10.1101/2021.01.06.425392v1
- Peer reviewed and published scientific report.
Zahradník, Jiří, Shir Marciano, Maya Shemesh, Eyal Zoler, Daniel Harari, Jeanne Chiaravalli, Björn Meyer, et al. 2021. “SARS-CoV-2 Variant Prediction and Antiviral Drug Design Are Enabled by RBD in Vitro Evolution.” Nature Microbiology 6 (9): 1188–98. https://doi.org/10.1038/s41564-021-00954-4. https://www.nature.com/articles/s41564-021-00954-4.