Researchers in the United States have demonstrated a potential new approach to treating infection with the new variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19).
The team used all-atom molecular dynamics to explore the mechanisms underlying the increased infectivity and spread of the recently emerged B.1.1.7 lineage that has been designated a variant of concern by Public Health England.
The researchers showed that a mutation within B.1.1.7 increases the virus’s ability to bind to host cells through hydrophobic interactions with the host cell receptor angiotensin-converting enzyme 2 (ACE2).
More specifically, a mutation called N501Y located in the receptor-binding domain (RBD) of the viral spike protein forms hydrophobic interactions with ACE2 residues Y41 and K353 to enhance RBD-ACE2 interfacial binding.
The team – from the Thomas J. Watson Research Center in New York and Neoland Biosciences in Massachusetts – also investigated how the N501Y mutation might enable the virus to evade attack by neutralizing antibodies.
Binquan Luan and colleagues suggest a modification to the therapeutic monoclonal neutralizing antibody CB6 that could increase its binding affinity for the spike RBD (sRBD).
“We expect that our work can help researchers design proper measures responding to this urgent virus mutation, such as adding a modified/new neutralizing antibody specifically targeted at this variant in the therapeutic antibody cocktail,” they write.
A pre-print version of the paper is available on the bioRxiv* server, while the article undergoes peer review.
The mechanism underlying the N501Y mutation is not yet clear
Since the first cases of SARS-CoV-2 were first identified in Wuhan, China late last year (2020), the unprecedented spread of the virus has led to more than 87.3 million cases of COVID-19 and more than 1.8 million deaths.
Recently, the World Health Organization identified an additional mutation – N501Y – in the SARS-CoV-2 D614G variant that has now emerged completely independently in the new B.1.1.7 lineage.
The researchers say the molecular mechanism underlying the characteristic N501Y mutation is not yet clear. However, high-throughput screening of all potential mutations in sRBD has predicted that the mutation can enhance binding between sRBD and ACE2.
“So far, the molecular mechanism of this enhanced binding is still elusive, which prevents us from assessing its effects on existing therapeutic antibodies,” writes Luan and colleagues.
Currently, neutralizing monoclonal antibodies (mAbs) represent a promising therapeutic and prophylactic approach to COVID-19, but how the N501Y mutation might affect the binding affinity of neutralizing antibodies for the sRBD remains unknown.
What did the researchers do?
Using all-atom molecular dynamics (MD), the researchers visualized ACE2-RBD binding at the atomic level. They investigated how the N501Y mutation can cause conformational changes for residues located at the ACE2-RBD interface.
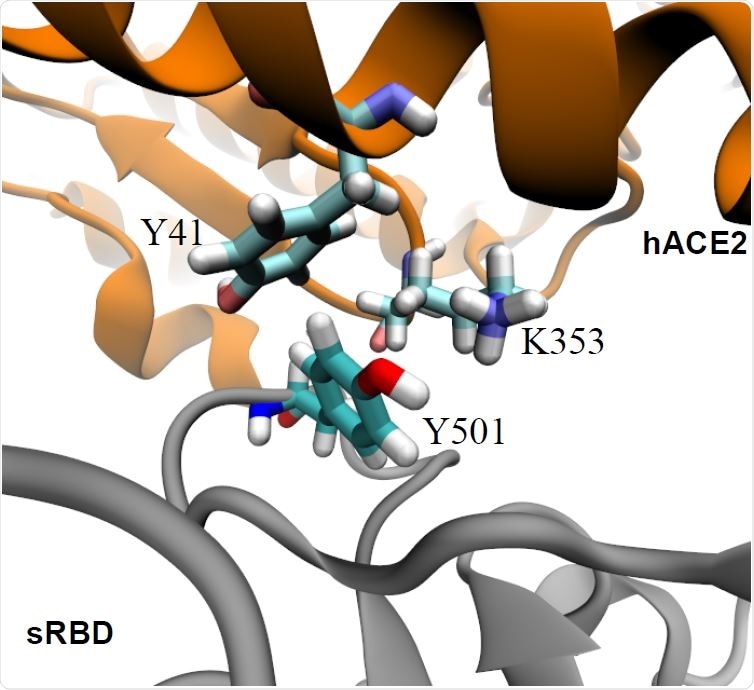
Enhanced interfacial coordinations between Y501 in sRBD and key residues (Y41 and K353) in hACE2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The team found that the wild-type spike protein N501, which is hydrophilic, is unfavorably coordinated with hydrophobic fragments in the Y41 and K353 residues of ACE2.
Luan and colleagues, therefore, hypothesized that these interfacial interactions would be improved with mutation of N501 into a hydrophobic residue.
Previous experimental screening of all potential sRBD mutations has consistently shown that mutating N501 into V, F, W, or Y can enhance sRBD-ACE2 binding.
“In the emergent SARS-CoV-2 variant, the presence of the N501Y mutation in sRBD indeed caused the disease to become more contagious, a consequence of the enhanced binding between the hACE2 and sRBD,” write the researchers.
To investigate the underlying molecular mechanism of the N501Y mutation, the team performed free energy perturbation (FEP) experiments to calculate the binding free energy difference for the N501Y mutation on the sRBD.
Effects of the N501Y mutation on the binding between the neutralizing mAb CB6 and sRBD. a) The complex of sRBD and the Fab (in CB6) equilibrated in MD simulation. The Fab comprises one heavy chain (fragment) and one light chain, colored in blue and orange respectively; the sRBD is in gray. The heavy (light) chain contains a variable region VH (VL) and a constant region CH (CL). b) Coordinations between N501 in sRBD and its surrounding residues (S30, R31 and Y32) in Fab, at the beginning of the FEP calculation. c) Coordinations between Y501 in sRBD and surrounding residues (S30, R31 and Y32) in Fab, at the end of the FEP calculation.
This showed that the mutation increased the binding affinity between ACE2 and sRBD by about 0.81 kcal/mol. The hydrophobic pocket formed by residues Y41 and K353 in ACE2 fitted well with Y501 in the sRBD.
“Therefore, Y501 in sRBD coordinates very well with both Y41 and K353 in hACE2,” say the researchers.
What about the effects on neutralizing antibodies?
Since many mAbs bind to the site in sRBD that overlaps with the binding site of ACE2, the team investigated how the N501Y mutation might affect the binding affinity of one such mAB called CB6.
Using the previous MD simulation, the team obtained the bound state for the complex of sRBD and the CB6 Fab region.
Using FEP calculations, the team found that the N501Y mutation can weaken the binding between sRBD and this Fab region.
Further analysis of the atomic coordination between N501 in sRBD and surrounding residues in the Fab region showed that the Fab residue R31 forms a direct interaction with N501. However, the hydrophobic Y501 repels the charged guanidino group in R31.
Modifying CB6 to accommodate the mutation
“As a biologics drug, the CB6 can be modified accordingly to accommodate the N501Y mutation in sRBD,” says Luan and colleagues.
The team, therefore, recommends mutating residue 31 in the light chain of the CB6 Fab region from R into hydrophobic F, Y or W.
This might “stabilize and improve the interfacial binding between sRBD and the modified CB6 antibody,” say the researchers.
“This modified mAB specifically targeting the SARS-CoV-2 variant might be added into the antibody cocktail to treat all COVID-19 patients,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources