The novel SARS-CoV-2 virus is the agent responsible for the coronavirus disease 2019 (COVID-19) pandemic that continues to sweep the globe posing an unprecedented threat to global health and the worldwide economy.
A significant challenge for viruses such as SARS-CoV-2 is the specific and efficient packaging of a large genome into a relatively small capsid while excluding viral subgenomic fragments and cellular nucleic acids.
How these viruses select the genome from the sub-genomic RNAs that the virus and host transcriptome generate and manage to compress it into a virion is not yet clear.
Now, using a coarse-grained polymer model, Amy Gladfelter and colleagues have shown that a process called phase separation promotes conditions that favor singular packaging. However, the arrangement of phase-separation-promoting-sequences at the ends of the genome is crucial for genome compaction. The elements must also be positioned at both ends of the genome for cyclization to occur.
The teams say the models presented here may be applicable to other viruses and cellular systems that involve long nucleotide chains and proteins.
A pre-print version of the paper is available on the bioRxiv* server, while the article undergoes peer review.
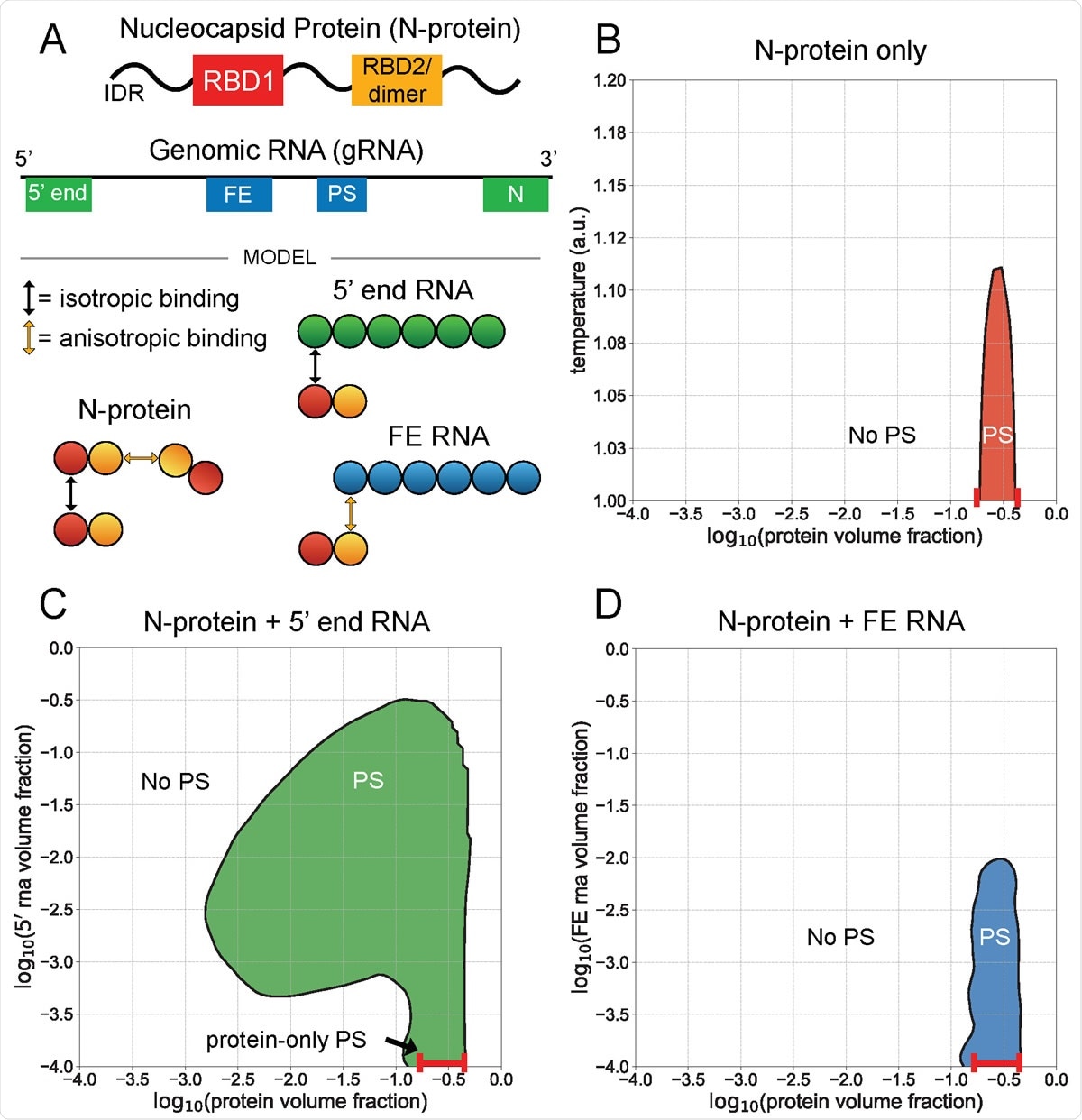
5¢ end and FE RNA with N-protein have opposing phase behavior. A) N-protein is represented as a two-bead chain, with the first bead participating in isotropic homotypic interactions, and the second bead participating in anisotropic homotypic interactions. Both 5¢ end and FE RNA segments are roughly three times larger than N-protein and are represented by six beads each. N-protein interacts with all 5¢ end RNA beads via isotropic binding with its first bead, and it interacts with all FE RNA beads via anisotropic binding with its second bead. This interaction with FE competes with N-protein dimerization. B) N-protein phase separates (PS) in a narrow concentration and temperature range on its own. C) N-protein with 5¢ end RNA at temperature 1 a.u. phase separates across a wider concentration range than on its own. D) N protein with FE RNA at temperature 1 a.u. is solubilized at sufficiently high FE RNA concentrations.
Understanding coronavirus replication
The COVID-19 pandemic has inspired great research efforts to understand the mechanisms underlying coronavirus replication.
Viruses must selectively package their large genomes into a relatively small capsid while excluding host nucleic acids and viral sub-genomic fragments.
Studies have recently demonstrated that viral proteins required for genome packaging can undergo a process called liquid-liquid phase separation (LLPS), where one or two proteins that bind viral nucleic acid condense with the genome.
In the case of SARS-CoV-2, the genomic RNA can condense with the nucleocapsid (N) protein, a protein that is essential for genome packaging in many viruses.
Importantly, some regions of the genomic RNA drive condensation of the N protein, while others solubilize it.
Previous work by Gladfelter and colleagues found that the N protein undergoes LLPS in an RNA sequence-specific manner within different regions of the genome.
“Remarkably, RNAs of the same length can either promote or limit phase separation depending on their sequences,” say the researchers.
They found that the LLPS- promoting sequences are in the 5¢ and 3¢ ends of the genome. This led the team to hypothesize that phase separation could be relevant to packaging since these sequences occur specifically on the whole genome and would not occur together on sub-genomic or host nucleic acids.
What did the current study involve?
The researchers used a coarse-grained polymer model to explore how the spatial patterning of LLPS-promoting or LLPS-inhibiting RNA regions in the genome might promote the specific packaging of a single genome.
Firstly, the team analyzed portions of the SARS-CoV-2 genome that exhibit opposing phase behavior once mixed with the N protein.
The researchers showed that while the 5¢ and 3¢ ends of the genome promote phase separation, the frameshifting element (FE) and central regions solubilize N protein.
Next, the team examined the spatial patterning of these opposing elements within a full genome model and the effects this patterning had on phase separation and single genome packaging, compaction and cyclization.
The model revealed that the most optimal arrangement for genome packaging and compaction was the localization of LLPS-promoting elements to the 5¢ and 3¢ ends of the genome and that this localization is necessary for cyclization.
Future applications of the model
The researchers say the model can inform future studies investigating spatial patterning of genomic features that are experimentally intractable because of the large genome size.
“We hypothesize that this model should be generalizable to multiple viruses as well as cellular organelles,” they write.
“Components from several viruses have been shown to undergo phase separation, raising the possibility that spatial patterning of specific LLPS-promoting RNA or DNA sequences may have evolved to promote optimal genome packaging in other viruses in addition to SARS-CoV-2,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources