The COVID-19 pandemic is still going strong a year after the novel coronavirus originated in Wuhan, China, with the virus mutating into more infectious variants. A fast and accurate virus detection strategy is very crucial to reduce the rate of transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen responsible for COVID-19. The global scientific research community responded to this unprecedented demand in diagnostic testing by developing several detection platforms, the most sensitive of which detect the viral RNA.
The current gold standard in testing - reverse transcriptase-polymerase chain reaction (RT-PCR) - is a 2-step assay that takes over 60 minutes to test one sample. Reverse transcriptase is an enzyme that converts viral RNA to complementary DNA (cDNA), which takes nearly 30 minutes. The cDNA is then amplified using a quantitative PCR (qPCR), and the amplified DNA is detected with the help of a fluorescent dye, which can take about an hour.
Reducing detection times and increasing sample throughput is crucial in bringing down the rate of spread of the virus. The research community has discussed several new approaches that help reduce assay times for detecting SARS-CoV-2 over the past one year.
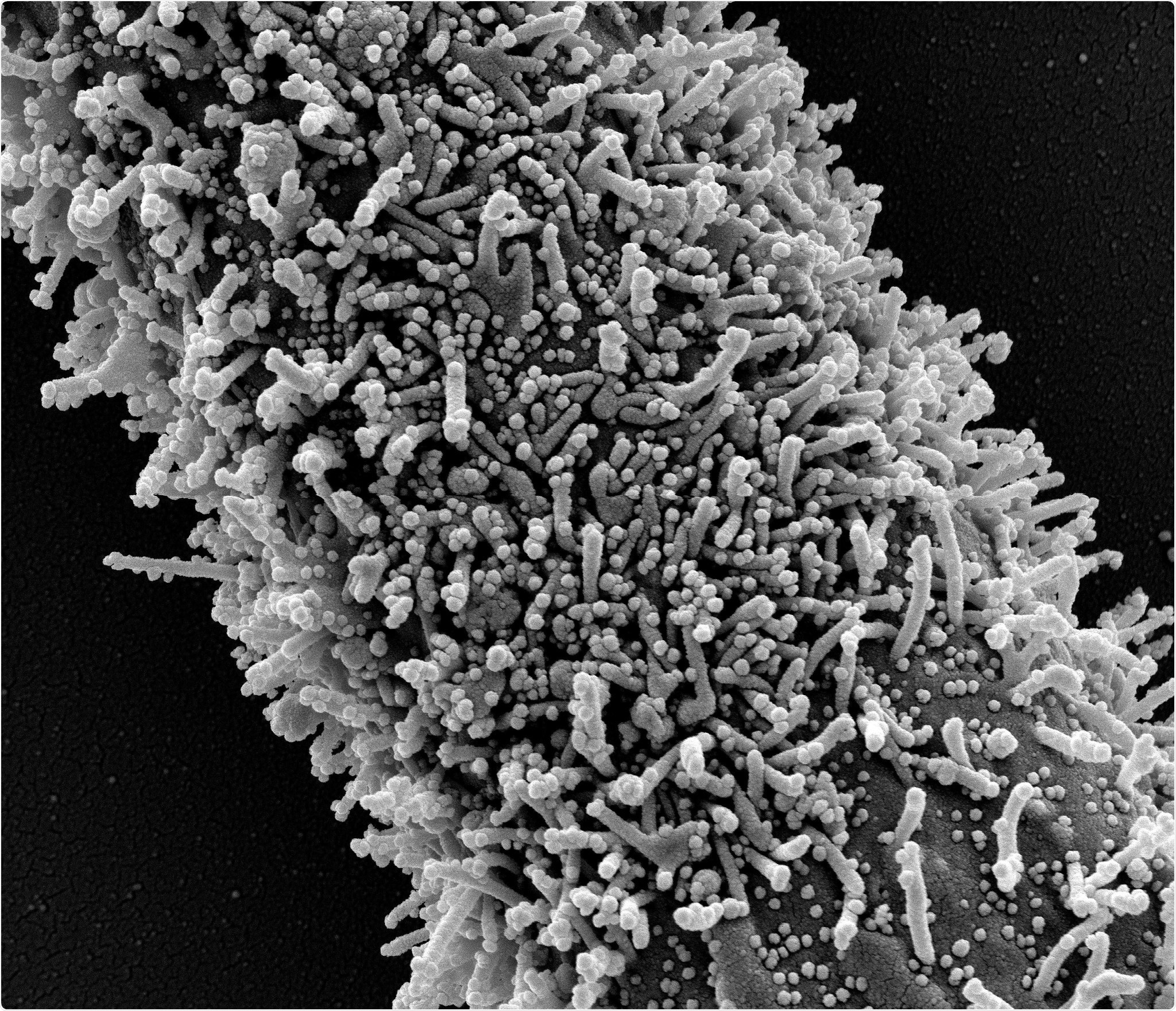
A single elongated CCL-81 cell heavily infected with SARS-CoV-2 virus particles. The small spherical structures in the image are SARS-CoV-2 virus particles. The rod-shaped protrusions from the cells are cell projections or pseudopodium. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
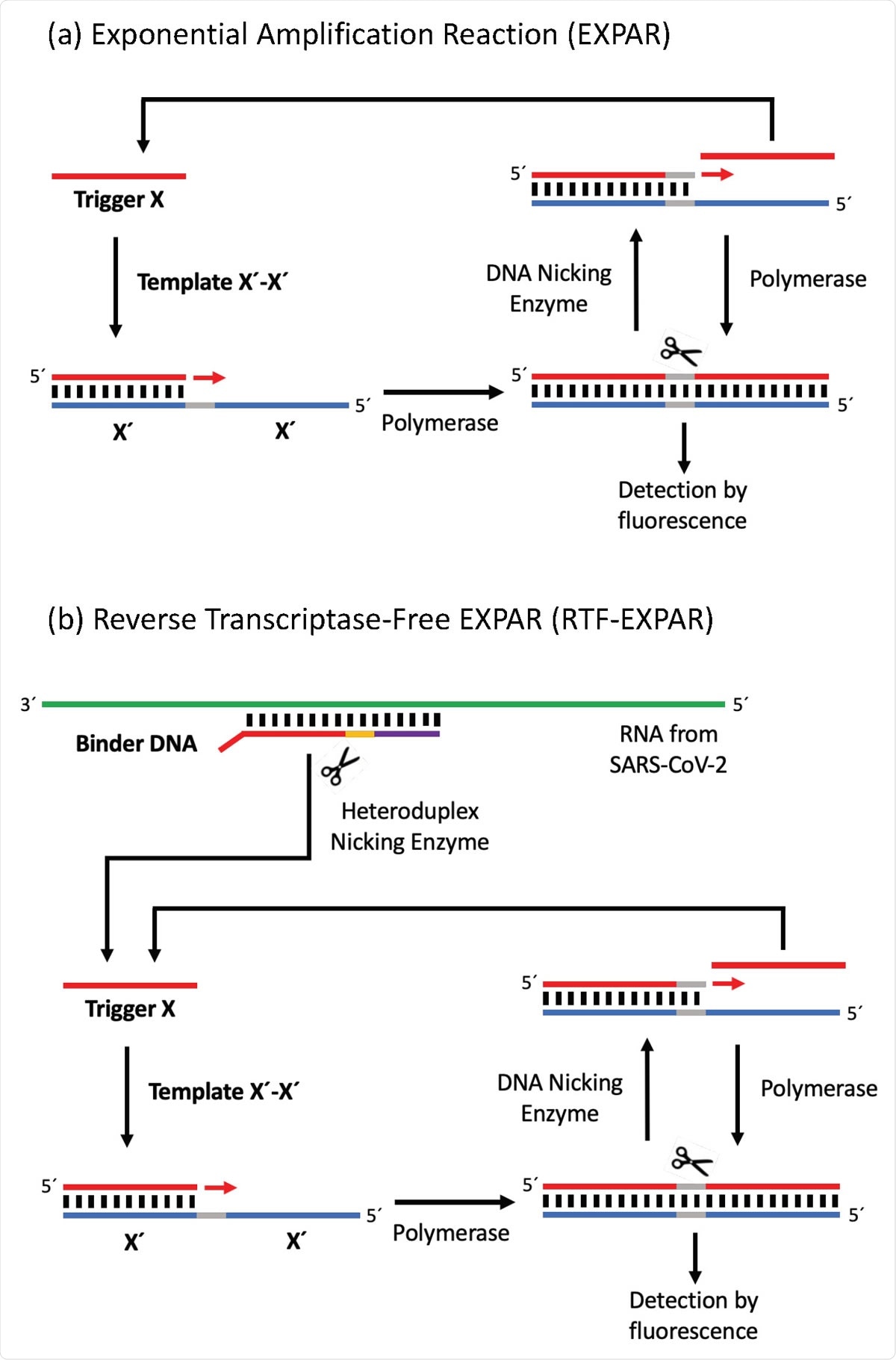
Reverse transcriptase-free isothermal SARS-CoV-2 detection
In a recent medRxiv* preprint research paper, researchers from the UK reported a rapid isothermal method for SARS-CoV-2 detection. The researchers used a novel approach for converting RNA into cDNA without using the reverse transcriptase enzyme. This new technique rapidly amplifies the DNA using the exponential amplification reaction (EXPAR).
Since the new reverse transcriptase-free (RTF-EXPAR) approach combines the conversion of RNA to DNA and amplification of the cDNA using EXPAR in one single step, it can SARS-CoV-2 detection in a sample in less than 5 minutes.
How does the EXPAR approach work?
In the EXPAR technique, the amplification of cDNA happens at a single temperature, which avoids time-consuming heating and cooling steps. Also, the amplicon is relatively small - typically about 15-20 bases long - compared to both PCR and loop-mediated isothermal AMPlification (LAMP) techniques. Thus, once triggered, EXPAR can produce up to 108 strands of DNA in a few minutes. Similar to the RT-PCR COVID-19 assay, duplex formation is monitored in EXPAR spectroscopically with the help of a fluorescent intercalating dye such as SYBR Green.
RTF-EXPAR assay data for SARS-CoV-2 RNA detection (72.7 copies/µL, n = 3), showing: (a) the mean time for the amplification reaction only and (b) the mean total assay time from RNA sample to signal. Each run time was calculated to be the point at which the fluorescence signal was greater than 10 standard deviations from the baseline signal (10-sigma time). Error bars in datasets are the standard deviations of the 10-sigma time. Signals observed for negative samples at >10 min are attributed to amplification arising from non-specific interactions.
“A crucial element to developing a successful EXPAR assay is the identification of optimal nucleotide sequences in the target genome.”
The researchers applied this EXPAR approach in a two-stage process. First, they performed enzymatic digestion at 50°C for 5 minutes of Binder DNA (1 µM) in the presence of a SARS-CoV-2 RNA (72.7 copies/µL) patient sample. Then they added this solution to the EXPAR reagent mix for amplifying the DNA obtained. Performed in triplicate, this step offered an amplification time of 3.17 ± 0.24 minutes.
RTF-EXPAR assay could be modified for use in the detection of other RNA viruses
To summarize, the researchers used a new reverse transcriptase-free isothermal amplification technique named RTF-EXPAR to demonstrated successful SARS-CoV-2 RNA detection with a total assay time of under 5 minutes. This approach is not only much faster than the RT-PCR assay, which needs at least 60 minutes to test one sample but also outperforms the LAMP and lateral flow antigen techniques.
“This time is not only much faster than RT-PCR (assay time of at least 60 minutes) but also outperforms LAMP and 30-minute lateral flow antigen tests in current deployment.”
According to the authors, RTF-EXPAR should be fully compatible and deployment-ready for use on the same equipment currently being used for RT-PCR COVID-19 assays. Moreover, the speed and simplicity of the essay make it possible to customize this method for use in the detection of a range of other infectious diseases that are caused by RNA-based viruses such as Ebola and RSV.
“In conclusion, through the use of a new reverse transcriptase-free isothermal amplification method, RTF-EXPAR, involving a DNA-selective restriction endonuclease, we have demonstrated the successful detection of SARS-CoV-2 RNA in a total assay time of less than 5 minutes.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Sub-5-minute Detection of SARS-CoV-2 RNA using a Reverse Transcriptase-Free Exponential Amplification Reaction, RTF-EXPAR, Jake G. Carter, Lorea Orueta Iturbe, Jean-Louis H. A. Duprey, Ian R. Carter, Craig D. Southern, Marium Rana, Andrew Bosworth, Andrew D. Beggs, Matthew R. Hicks, James H. R. Tucker, Timothy R. Dafforn, medRxiv 2020.12.31.20248236; doi: https://doi.org/10.1101/2020.12.31.20248236, https://www.medrxiv.org/content/10.1101/2020.12.31.20248236v1
- Peer reviewed and published scientific report.
Carter, Jake G., Lorea Orueta Iturbe, Jean-Louis H. A. Duprey, Ian R. Carter, Craig D. Southern, Marium Rana, Celina M. Whalley, et al. 2021. “Ultrarapid Detection of SARS-CoV-2 RNA Using a Reverse Transcription-Free Exponential Amplification Reaction, RTF-EXPAR.” Proceedings of the National Academy of Sciences of the United States of America 118 (35): e2100347118. https://doi.org/10.1073/pnas.2100347118. https://www.pnas.org/doi/full/10.1073/pnas.2100347118.