Drugs that target these “less variable” regions are more likely to be effective against future variants of SARS-CoV-2 because they remain unchanged or “conserved” as the virus mutates and evolves.
Daria Mochly-Rosen and colleagues identified the regions using a database of almost 190,000 individual virus sequences isolated from patients across the globe, thereby reflecting the natural evolution of the virus.
The team focused on data for the viral spike protein – the main structure the virus uses to gain access to host cells. The receptor-binding domain (RBD) of the spike protein attaches to the human receptor angiotensin-converting enzyme 2 (ACE2) as the initial step in the infection process.
Currently, much of the effort to develop prophylactic agents as treatments for COVID-19 is focused on targeting this spike protein to block its interaction with ACE2.
Now, Mochly-Rosen and colleagues suggest that it may be beneficial to develop passive and active vaccines that target the RBD, rather than the entire spike protein.
They also suggest that small molecules mimicking a region called the linoleic acid (LA)-binding site may be beneficial. In addition, they say that drugs targeting a relatively conserved and exposed region on the protein’s surface (spanning residues 541-612) may be of prophylactic benefit.
A pre-print version of the paper is available on the bioRxiv* server while the article undergoes peer review.
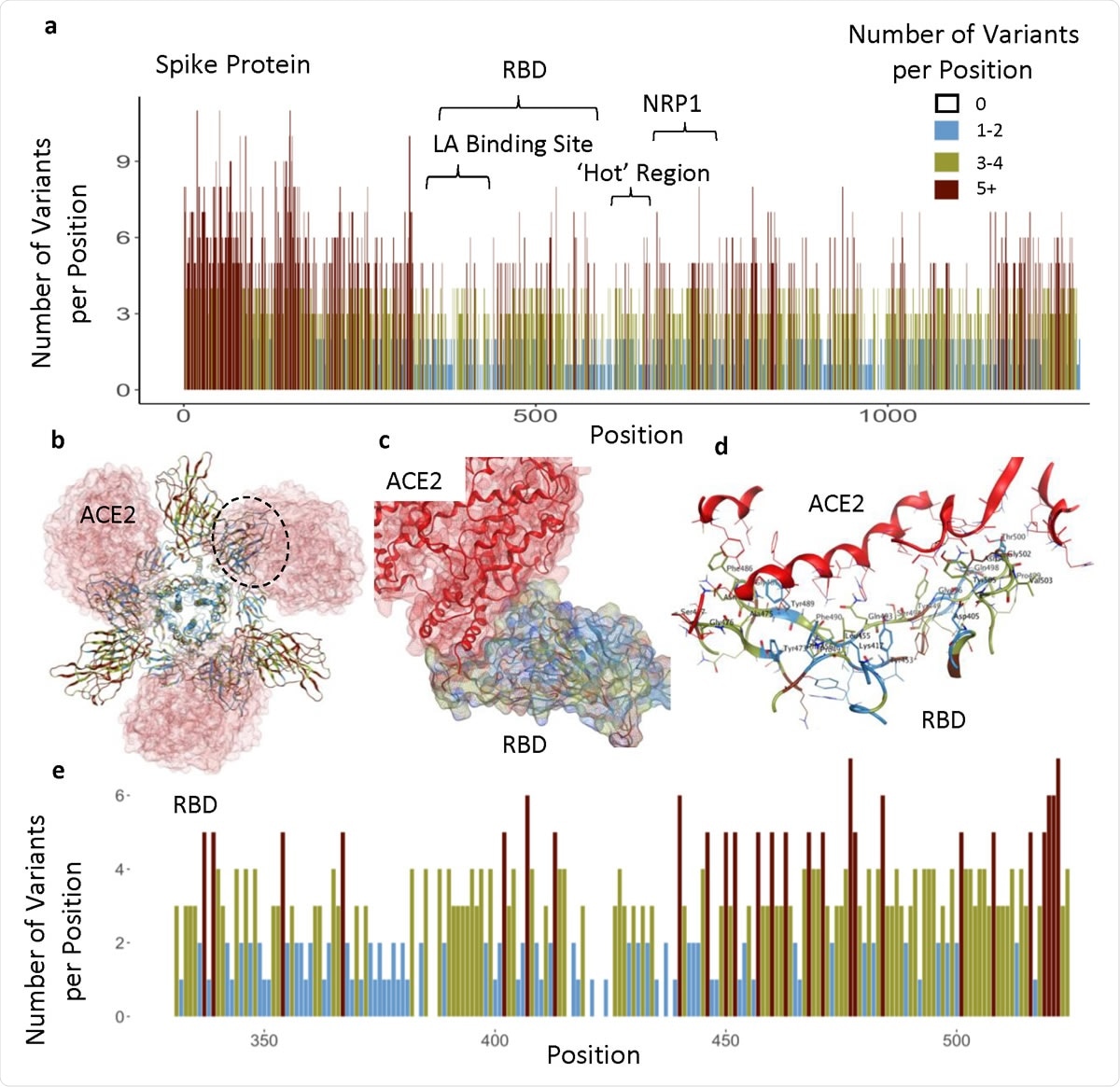
Functional regions in S protein and the RBD-ACE2 interaction site. a) The number of variants per position across entire sequence of S protein, highlighting specific functional regions. b) Spike homotrimer with ribbons colored according to legend, bound to ACE2 (red). Black dotted outline shown in c. c) RBD-ACE2 interface. d) RBD-ACE2 interface highlighting residues in RBD within 4.5Å from ACE2. e) The number of variants per position across RBD domain.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The SARS-CoV-2 infection process
As part of the SARS-CoV-2 infection process, proteases cleave the spike protein into two subunits. Subunit 1 (S1) contains the RBD in either a closed (inactive) conformation that does not enable viral binding or an open (active) conformation that does enable binding. When the RBD is in the open conformation, subunit (S2) enables the subsequent fusion of the virus to the host cell membrane.
Prophylaxis approaches involve preventing proteolytic cleavage of S1, competing with RBD binding, generating antibodies against the spike protein or RBD (passive vaccines), and triggering an immune response to the spike protein (active vaccines).
Aside from the RBD, other regions such as the spike trimer interface and the LA-binding site are likely key to maintaining SARS-CoV-2 structural integrity, entry, and transmission, say Mochly-Rosen and colleagues.
The researchers hypothesized that these regions of the spike protein would be more conserved and contain fewer variants than other regions.
What did the researchers do?
To identify regions in the spike protein that are the least variable, the team used the spike protein database created by the Singapore Bioinformatics Institute.
As of November 11th, 2020, the database included 189,704 individual sequences of the natural virus isolated from many patients worldwide.
Every variant observed in the spike protein was uploaded to the software tool PROVEAN and compared with the spike protein amino acid sequence from the Wuhan reference (EPI_ISL_402124) obtained in February 2020.
What did they find?
Compared with the reference sequence, the spike protein (comprising 1,273 amino acids) was found to have 4,517 variants.
Each amino acid position in the protein sequence harbored an average of four variants. However, some regions had ten variants at each position, while others had none. Regions that had no more than two variants per amino acid position were more prevalent in the structurally critical trimer interface (38%), the LA-binding site (65%), and the RBD (37%).
The researchers say the trimer interface is less accessible and therefore unlikely to be targeted by drugs. However, the LA-binding site could be a potential target for therapeutics with small molecules that stabilize the spike protein in its closed, inactive conformation.
“The fatty acid-binding pocket in the inactive conformation of S [spike] protein is conserved in other coronaviruses and 82% of the variants among the 20 amino acids that make this pocket are predicted to have a neutral effect,” writes the team.
What about the RBD?
The study found that the RBD (amino acids 331-524) contains ten invariant amino acids, and the PROVEAN tool predicted that only 7% of RBD variants were predicted to be structurally or functionally damaging.
“Therefore, drugs targeting the RBD are expected to be effective prophylactics to most SARS-CoV-2 variants,” write the researchers.
The team also identified another less variable or so-called “hot” region spanning amino acid residues 541-612.
The researchers say that within this region, the amino acid at position 564 has been proposed to serve as a “latch” that stabilizes the spike protein in the closed conformation.
The team suggests that this hot region may be involved in membrane fusion.
“Determining the role of this invariable region is important, as it may be another Achilles heel to target for anti-SARS-CoV-2 treatment,” writes Mochly-Rosen and the team.
What are the study implications?
The researchers say the findings could guide the development of effective prophylactic agents to arrest the spread of the COVID-19 pandemic.
“Our data suggest that it may be beneficial to develop passive and active vaccines that target the RBD, instead of the entire S protein, as well as small molecules that lock the closed conformation, such as those mimicking the LA,” they write.
“Finally, drugs and antibodies targeting region 541-612, a relatively conserved and exposed region on the protein’s surface, may also have prophylactic benefit,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources