A team of scientists from the University of British Columbia, Canada, and the University of Pittsburgh Medical School, USA, recently conducted structural analysis of a newly emerged UK variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In their analyses, they observed that the N501Y mutation in the receptor-binding domain (RBD) of the virus’s spike protein increases the receptor binding efficiency of the virus, thus making it more infectious. Most importantly, however, the study reveals that the mutated variant of SARS-CoV-2 can still be neutralized by specific antibodies, and that the N501Y mutation cannot influence the epitope binding ability of strong neutralizing antibodies. The study is currently available on the bioRxiv* preprint server.
_-_1200_width-2.jpg)

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Since its emergence in December 2019, SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), has already undergone more than 13,000 mutations. Of these mutations, D614G occurring in the receptor-binding domain (RBD) of the viral spike protein has been shown to increase the transmissibility of the virus without significantly affecting its pathogenicity. Recently, a newer variant of SARS-CoV-2 with multiple spike protein mutations has emerged in the UK. In this newer variant, the asparagine to tyrosine mutation at residue 501 in the spike RBD (N501Y mutation) is expected to increase the infectivity of SARS-CoV-2. Specifically, X-ray crystallography and cryo-electron microscopy (cryo-EM) structural studies have shown that N501Y mutation may increase the binding affinity of spike RBD to angiotensin-converting enzyme 2 (ACE2), thus increasing the viral infectivity.
Current study design
The current study was designed to investigate the impact of N501Y mutation on viral transmissibility/infectivity, viral entry, and antibody-mediated viral neutralization. The scientists conducted cryo-EM structural analysis of the SARS-CoV-2 spike protein with and without N501Y mutation, and investigated whether the mutation can affect the RBD epitope binding ability of a potent anti-SARS-CoV-2 neutralizing antibody, namely VH -Fc ab8.
Important observations
The cryo-EM analysis data have shown that N501Y mutation does not significantly alter the secondary or quaternary structure of the viral spike protein. By conducting structural analysis of the complex formed between the antibody and the spike protein without N501Y mutation, the scientists have observed that the residue 501 is located at the outer margin of the antibody footprint on the viral surface and that there is minimal interaction between the residue 501 and the antibody. Thus, it is less likely that a mutation in residue 501 will significantly affect the binding efficiency of the antibody.
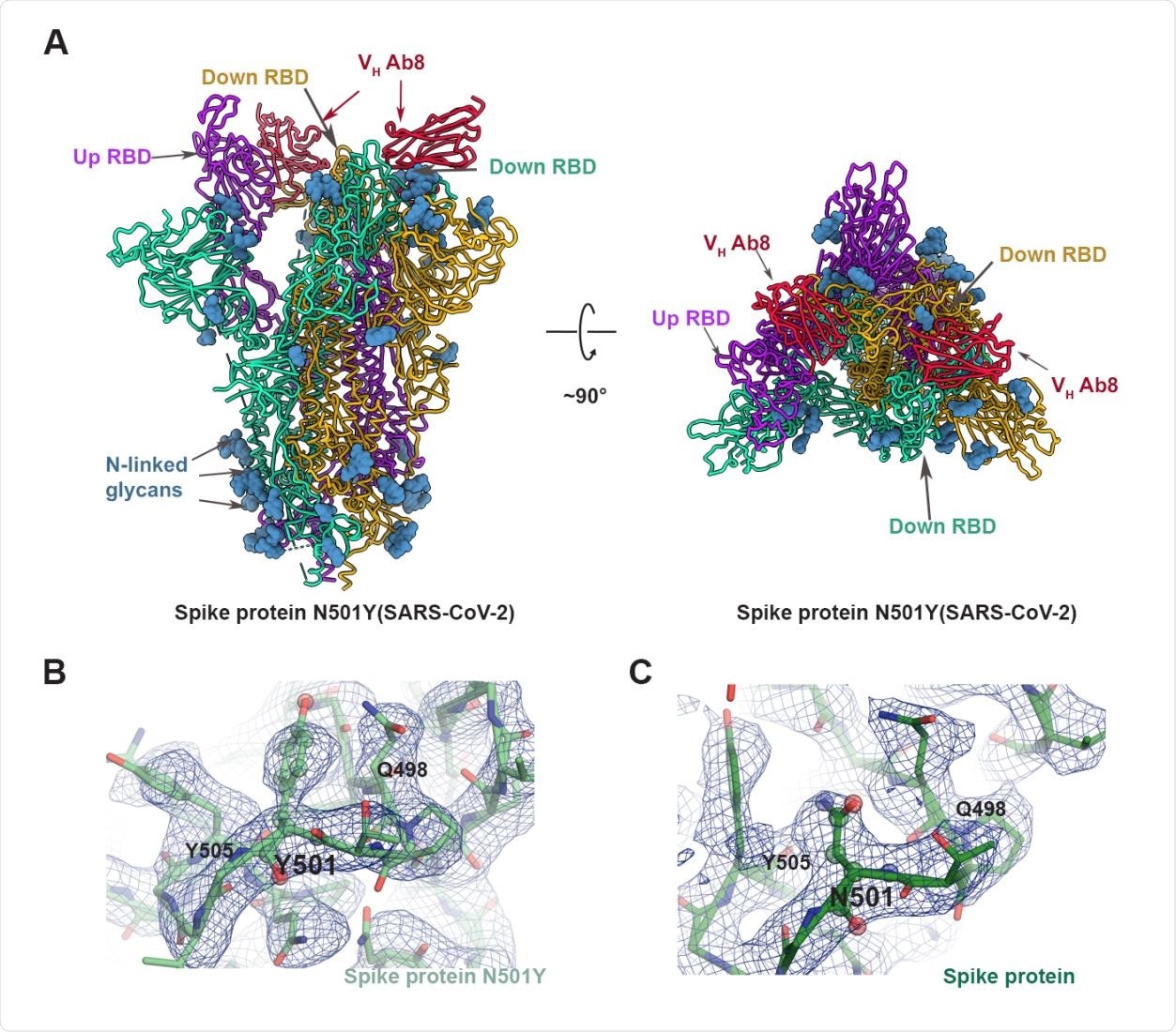
Structure of VH ab8 bound to spike protein trimer N501Y mutant. (A) Atomic model for the structures of the complex of VH ab8 with the N501Y mutant spike protein ectodomain. The structure has two RBDs in the down position with well-resolved densities for the bound VH ab8, with the third RBD in the up position, and poorly resolved VH ab8 density. (B, C) Views of the density map in the region near 501 for the N501Y mutant spike protein ectodomain (B) and the unmutated form (C).
Furthermore, the scientists have conducted structural analysis of the viral spike protein with N501Y mutation and observed that the mutation does not significantly affect the interaction between spike RBD and the antibody. However, because the tyrosine side chain is bulkier than the asparagine side chain, the N501Y mutation has been shown to cause minute local structural rearrangements.
Regarding the spike RBD and ACE2 interaction, the study findings demonstrate that the residue 501 is located at the outer boundary of the region required for the interaction between RBD and ACE2. By experimentally infecting ACE2-expressing cells with mutated or unmutated SARS-CoV-2, the scientists have observed that N501Y mutation significantly increases the viral ability to infect host cells. With further analysis, they have noticed that the increased infectivity of the mutated virus is due to improved binding efficiency of the spike RBD to ACE2.
By conducting neutralization experiments, they have observed that N501Y mutation does not affect the interaction between the antibody and the neutralizing epitopes in the RBD and that the antibody can neutralize both mutated and unmutated SARS-CoV-2 with similar efficiency. Moreover, the N501Y mutant has been found to compete for binding with the spike – ACE2 interaction in a dose-dependent manner.
Study significance
The study reveals that the newly emerged mutation (N501Y) in SARS-CoV-2 has significantly increased the viral infectivity, justifying the rapid global spread of the “UK variant of SARS-CoV-2”.
The most important observation is that despite improving the viral fitness in terms of increased infectivity, SARS-CoV-2 with N501Y mutation can still be efficiently neutralized by strong neutralizing antibodies. Thus, it can be expected that the COVID-19 vaccines that are currently in production can be effective in providing protection against the mutated viral variant.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources