The coronavirus disease 2019 (COVID-19) pandemic continues to ravage the globe. To date, over 91.62 million cases and more than 1.96 million deaths have been reported worldwide.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes COVID-19, is commonly thought to be a zoonotic disease that originated in bats and then subsequently jumped to humans. This is thought to have occurred via an as yet unidentified intermediate host that may have brought the virus to Wuhan City, China, where the pathogen was first detected in December 2019.
Now, a new study by based researchers at the University of Cambridge and the Pirbright Institute in the UK has identified key genetic changes in SARS-CoV-2 that may be responsible for the virus’s jump from bats to humans. The team also determined which animals contain cellular receptors that allow the virus to enter cells more effectively, zoning in on potential animals that acted as an intermediary host in facilitating SARS-CoV-2’s zoonosis.
The coronavirus pandemic
SARS-CoV-2 first emerged in late 2019 in China, and has since spread to 191 countries and territories. To date, it has caused more than 91.71 million cases and 1.96 million deaths.
Coronaviruses commonly circulate among bat species, and SARS-CoV-2 – a positive-strand RNA pathogen of the betacoronavirus family – is also thought to have originated in the Chiroptera species in China. Yet, whether the virus directly jumped to humans or through an intermediate host is still unclear. However, it has been established that the virus can infect companion animals, pets, wildlife, and livestock.
New variants of the virus are circulating across the globe, leading to successive waves of infections. Many governments in especially hard-hit countries – such as the UK, where a highly infectious new strain has been identified – have reimposed travel bans, restrictions, and lockdown orders to try and contain the virus's spread.
Bats to humans
The study, published in the journal PLOS Biology, used a combination of surrogate entry assays and live virus to demonstrate that aside from the human angiotensin-converting enzyme 2 (ACE2), the spike glycoprotein of SARS-CoV-2 (also known as the spike protein or S protein) has a wide range of host tropisms for mammalian ACE2 receptors. This is despite divergence in the amino acids at the spike protein’s receptor binding site.
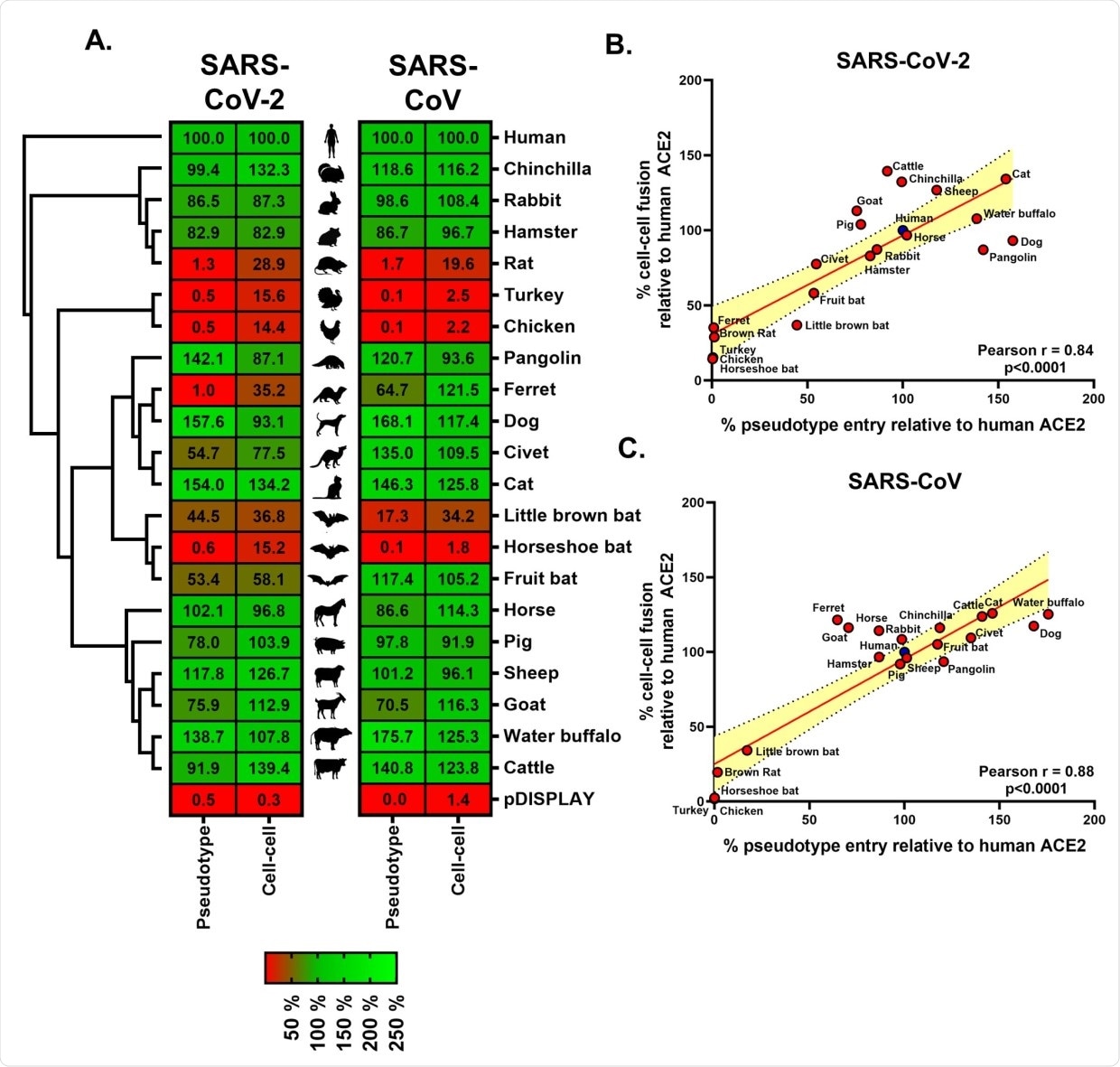
Receptor screening using surrogate entry assays identifies SARS-CoV-2 Spike as a pan-tropic viral attachment protein. (A) A heatmap illustrating the receptor usage profile of SARS-CoV-2 and SARS-CoV in pseudotype entry and cell–cell fusion assays with various mammalian and bird ACE2s. The data in each row are normalised to the signal seen for human ACE2 (top), with results representing the mean percentage calculated from 3 separate experiments performed on different days. A vector-only control (pDISPLAY) was added to demonstrate specificity. Mammalian and bird ACE2s are organised, top to bottom, based on their phylogenetic relationship (rectangular cladogram, left). The inter-experimental standard error of the mean for the pseudotype and cell–cell fusion assays ranged from 0.01% to 47.92% (median 10.73%) and 0.12% to 32.97% (median 5.43%), respectively. (B and C) For both SARS-CoV-2 and SARS-CoV, the respective cell–cell and pseudotype assay percentages for each ACE2 protein (relative to human ACE2) were plotted on an XY scatter graph, the Pearson correlation calculated and a linear line of regression fitted together with 95% confidence intervals. The data underlying this figure may be found in S1 Data. ACE2, angiotensin-converting enzyme 2; SARS-CoV, SARS Coronavirus; SARS-CoV-2, SARS Coronavirus 2.
Further, they found that the genetic adaptations identified were similar to those made by the severe acute respiratory syndrome coronavirus (SARS-CoV), which caused the 2002-2003 severe acute respiratory syndrome (SARS) outbreak. The study findings suggest that there may be a common mechanism by which coronaviruses mutate for them to jump from animal hosts to humans.
With relatives of SARS-CoV-2 – SARS-CoV and the Middle East Respiratory Syndrome coronavirus (MERS-CoV), which emerged in Saudi Arabia in 2012 – the animal reservoir is thought to be bats. Viral spillover into humans has been suspected and scientists believe this occurred via an intermediate host in both cases. The SARS-CoV is thought to come from chivets and MERS-CoV from camels.
Coronaviruses typically use their spike proteins to enter cells by binding with human cell surface receptors like the ACE2. These binding sites act like a lock and key, wherein the spike protein needs to be the right shape to fit into the receptors and facilitate viral entry. However, different species express receptors with varying shapes, which means that the virus’s spike proteins may bind with the receptors more tightly in certain animals than others.
![The SARS-CoV-2 binding site on ACE2 is highly variable. (A) A phylogenetic tree of ACE2 proteins assembled using the neighbor-joining method [51] conducted in MEGA7 (Temple University, USA) [52] with ambiguous positions removed. The tree is drawn to scale, and support was provided with 500 bootstraps. (B) Structure of human ACE2 ectodomain (green) in complex with the RBD of SARS-CoV-2 [10]. (C) Conservation of mammalian ACE2 amino acid residues, estimated from site-specific evolutionary rates [50], mapped onto the surface of the ACE2 ectodomain [10], and coloured from blue (divergent) to purple (conserved) and presented in 2 orientations. Inset depicts the SARS-CoV-2 binding region of ACE2 (outlined), with residues that contact the SARS-CoV-2 RBD highlighted [6]. (D) WebLogo (University of California, Berkeley, USA) [53] plots summarising the amino acid divergence within the mammalian and bird ACE2 sequences characterised in this study. The single letter amino acid (aa) code is used with the vertical height of the amino acid The SARS-CoV-2 binding site on ACE2 is highly variable. (A) A phylogenetic tree of ACE2 proteins assembled using the neighbor-joining method [51] conducted in MEGA7 (Temple University, USA) [52] with ambiguous positions removed. The tree is drawn to scale, and support was provided with 500 bootstraps. (B) Structure of human ACE2 ectodomain (green) in complex with the RBD of SARS-CoV-2 [10]. (C) Conservation of mammalian ACE2 amino acid residues, estimated from site-specific evolutionary rates [50], mapped onto the surface of the ACE2 ectodomain [10], and coloured from blue (divergent) to purple (conserved) and presented in 2 orientations. Inset depicts the SARS-CoV-2 binding region of ACE2 (outlined), with residues that contact the SARS-CoV-2 RBD highlighted [6]. (D) WebLogo (University of California, Berkeley, USA) [53] plots summarising the amino acid divergence within the mammalian and bird ACE2 sequences characterised in this study. The single letter amino acid (aa) code is used with the vertical height of the amino acid](https://www.news-medical.net/image-handler/picture/2021/1/journal.pbio_2_-_1200_width.jpg)
The SARS-CoV-2 binding site on ACE2 is highly variable. (A) A phylogenetic tree of ACE2 proteins assembled using the neighbor-joining method [51] conducted in MEGA7 (Temple University, USA) [52] with ambiguous positions removed. The tree is drawn to scale, and support was provided with 500 bootstraps. (B) Structure of human ACE2 ectodomain (green) in complex with the RBD of SARS-CoV-2 [10]. (C) Conservation of mammalian ACE2 amino acid residues, estimated from site-specific evolutionary rates [50], mapped onto the surface of the ACE2 ectodomain [10], and coloured from blue (divergent) to purple (conserved) and presented in 2 orientations. Inset depicts the SARS-CoV-2 binding region of ACE2 (outlined), with residues that contact the SARS-CoV-2 RBD highlighted [6]. (D) WebLogo (University of California, Berkeley, USA) [53] plots summarising the amino acid divergence within the mammalian and bird ACE2 sequences characterised in this study. The single letter amino acid (aa) code is used with the vertical height of the amino acid.
Identifying the animal reservoir of SARS-CoV-2 and any intermediate hosts may help provide insights into how, where, and when the virus emerged and was eventually passed on to humans. It may also inform the development of preventative measures to ensure this form of zoonosis does not recur.
RaTG13 virus
In the SARS outbreak, scientists identified closely related isolates in both bats and civets, where the virus is thought to have emerged. In the current outbreak, however, further studies are still needed to determine the exact source of the virus and the intermediate host.
The SARS-CoV-2 virus has the sequence of a related bat coronavirus known as RaTG13, which shares 96 percent similarity to the genome of SARS-CoV-2.
To assess if the differences between SARS-CoV-2 and RaTG13 were involved in the adaptation of the virus to humans, the researchers interchanged the regions and observed how well the spike proteins bound with the human ACE2 receptors.
The study findings showed that the spike proteins of SARS-CoV-2 that contains RaTG13 regions could not bind to human ACE2 receptors effectively. On the other hand, RaTG13 spikes containing SARS-CoV-2 regions could bind more effectively to human receptors. The findings showed that similar changes in the spike protein happened historically, which may have played an essential role in the viral spillover.
Animal infection
Apart from the study findings, the researchers also found that dog, cattle, and cat ACE2 receptors have the strongest interactors with the SARS-CoV-2 spike protein. This means that these animals are at a higher risk of being infected with SARS-CoV-2.
As we saw with the outbreaks in Danish mink farms last year, it’s essential to understand which animals can be infected by SARS-CoV-2 and how mutations in the viral spike protein change its ability to infect different species,”
Dr. Stephen Graham in the University of Cambridge’s Department of Pathology
However, further research is needed to establish if these animals can acquire infection and if they could act as animal reservoirs.
Journal reference:
- Conceicao, C., Thakur, N., Humana, S., Kelly, J., Logan, L., et al. (2021). The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. PLOS BIOLOGY. https://doi.org/10.1371/journal.pbio.3001016, https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001016