Antibodies with heavy chain regions derived from a gene called VH1-2 constitute some of the most potent neutralizing antibodies against SARS-CoV-2 identified to date.
Now, Zizhang Sheng and colleagues have identified shared genetic and structural signatures within a class of VH1-2 antibodies that could improve efforts to elicit such antibody classes through vaccination.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
“Multi-donor” antibody classes show potent neutralizing activity against viruses
Studies have revealed prominent classes of similar neutralizing antibodies (nAbs) that frequently arise in response to viral infection or vaccination.
Such “multi-donor” antibody classes exhibit potent, broad neutralizing activity and frequently occur in the human antibody repertoire.
“A prominent mode of rational vaccine design – lineage-based vaccine design – endeavors to elicit such antibody classes by vaccination and this approach has recently entered clinical assessment,” says Sheng and colleagues.
The race to develop vaccines against SARS-CoV-2
Since the COVID-19 outbreak began in Wuhan, China, late last year, researchers have been racing to develop effective vaccines that could bring an end to the global public health crisis.
In the case of SARS-CoV-2, nAbs primarily target the viral spike protein – the structure that binds the host cell receptor angiotensin-converting enzyme 2 (ACE2) as the initial step in the infection process.
The spike protein binds to ACE2 via a region called the receptor-binding domain (RBD). This highly flexible RBD can adopt either an “up” (open) confirmation that is capable of interacting with ACE2 or a “down” (closed) conformation that is not.
Many nAbs have been characterized that bind to epitopes on either conformation and neutralize SARS-CoV-2 by competing with ACE2 for RBD binding or locking the RBD in the down conformation.
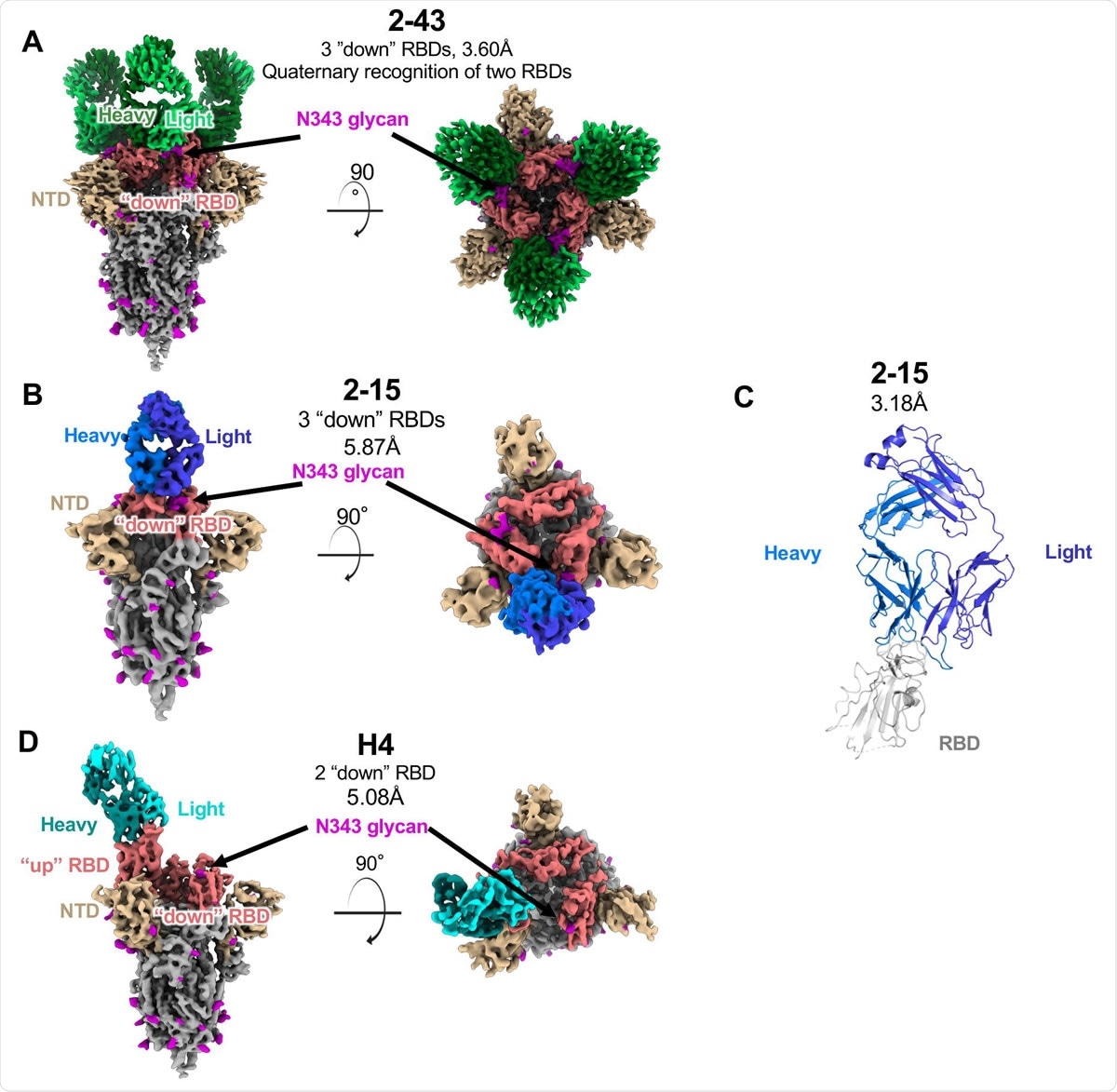
Structures of three SARS-CoV-2 neutralizing VH1-2 antibodies reveal both 'RBD-down' and 'RBD-up' modes of spike recognition. (A) Side and top views of three 2-43 antibody Fabs bound to prefusion SARS-COV-2 spike in the closed state. Color schemes are: 2-43 green; RBD, salmon; NTD, yellow; N-linked glycans, magenta; other spike regions, gray. (B) Side and top views of one 2-15 antibody Fabin complex with prefusion SARS-COV-2 spike in the closed state. 2-15 is colored blue. (C) Overview of the crystal structure of 2-15 in complex with SARS-CoV-2 RBD. Residue segments missing in the structure are shown as dashed lines. (D) Side and top views of one H4 Fab recognizing an "up" RBD on prefusion SARS-COV-2 spike. H4 is colored cyan.
Characterizing genetic features of these nAbs
Characterization of the SARS-CoV-2 nAbs so far identified has revealed enrichment of the antibody variable genes VH3-53, VH1-2, VH1-69, VH3-66, VH1-58, and VH3-30.
Structural characterization of the various nAbs has revealed two separate RBD-targeted classes that are derived from the similar VH3-53 and VH3-66 genes. One class that is characterized by a heavy chain complementarity-determining region 3 (CDRH3) of 15 amino acids or shorter targets RBD in the up conformation.
The other class, which has a longer CDRH3, recognizes a similar region of RBD, suggesting that CDRH3 is critical for the classification of VH3-53/-66 antibodies.
More about classes derived from the VH1-2 gene?
“Antibodies with heavy chains that derive from the VH1-2 gene constitute some of the most potent SARS-CoV-2-neutralizing antibodies yet identified,” writes Sheng and the team.
However, whether these antibodies form gene-restricted classes that represent shared effective antibody responses remains unclear.
Currently, three VH1-2 potent nAbs (2-4, S2M11, and C121) have been shown to exhibit similar modes of RBD recognition but different recognition of quaternary epitopes.
“It is thus still unclear whether RBD-targeting VH1-2 antibodies form a gene-restricted class,” say the researchers. However, “if so, what are the key genetic and structural signatures and determinants of neutralization potency?” they ask.
What did the team do?
The researchers determined the structures of three VH1-2-derived antibodies (2-15, 2-43, and H4) in complex with the SARS-CoV-2 spike protein.
All three of the antibodies used VH1-2-encoded motifs to recognize the RBD, with a somatic hypermutation (N531) in the heavy chain enhancing binding and light chain tyrosines recognizing the RBD residue F486.
Despite these similarities, VH1-2-antibody class members bound both RBD-up and RBD-down conformations, with a subset using elongated CDRH3 to recognize neighboring RBD – a strong quaternary interaction that is critical for locking RBDs in the down conformation.
What did the authors conclude?
The team says the findings show that the VH1-2 antibody class uses two modules for spike recognition, with the VH1-2 gene-encoded module used to recognize the RBD and the CDRH3 module used for quaternary recognition.
“Thus, we define a multi-donor VH1-2 antibody class, members of which can achieve very high neutralization potency, which is prevalent in human responses to SARS-CoV-2,” write the researchers.
“The shared genetic and structural signatures inform strategies to improve members of the VH1-2 antibody class,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Sheng R, et al. Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class. bioRxiv, 2020. doi: https://doi.org/10.1101/2021.01.11.426218, https://www.biorxiv.org/content/10.1101/2021.01.11.426218v1
- Peer reviewed and published scientific report.
Rapp, Micah, Yicheng Guo, Eswar R. Reddem, Jian Yu, Lihong Liu, Pengfei Wang, Gabriele Cerutti, et al. 2021. “Modular Basis for Potent SARS-CoV-2 Neutralization by a Prevalent VH1-2-Derived Antibody Class.” Cell Reports 35 (1): 108950. https://doi.org/10.1016/j.celrep.2021.108950. https://www.cell.com/cell-reports/fulltext/S2211-1247(21)00264-3.