As the coronavirus disease 2019 (COVID-19) pandemic continues to infect people in their hundreds of thousands around the world, a new preprint paper that appeared on the bioRxiv* server recently describes a panel of novel tests for genomic (g) and subgenomic (sg) mRNAs generated by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Most assays in use at present are either qualitative or semi-quantitative, and cannot evaluate the transcription molecules in detail. Detailed mapping of viral transcripts could help understand viral replication and immune evasion.
During this process, nine sgRNAs are also formed using a template switch to feed a leader sequence of ~70 nucleotides in the 5’ untranslated region (UTR), terminated with a short TRS at the 3’ end, to the sequence that awaits transcription, on one of the genes in a third of the genome at the 3’ end. The leader sequence TRS regulates the transcription of the sgRNAs and may allow differential expression of certain viral genes.
The 5’ UTR sequences may encode resistance to nsp1-mediated cleavage. Nsp1 may inhibit host gene expression by promoting the breakdown of host mRNA. The rapid build-up of sgRNAs in the host cell may enhance the severity of the disease. For positive-sense RNA viruses, sgRNAs act as regulators of virion production following translation of viral proteins, and also facilitate the translation of virulence or structural factors.
Genes targeted in the current study
The current study aimed to map various viral genes expressed in genomic or sgRNA molecules in a quantitative fashion, using a panel of seven ddPCR assays. The researchers selected the genes encoding the main protease Nsp5, RNA-dependent RNA polymerase (RdRp-Nsp12), four major proteins, namely, the spike (S), membrane (M), envelope (E) and nucleocapsid (N) protein, which have essential roles to play in SARS-CoV-2. For the spike protein gene, they used a primer set targeting the short polybasic cleavage site (S-PBCS) that is cleaved to the S1 and S2, the site that is novel to SARS-CoV-2 and that may enhance its infectivity in humans.
Sensitivity and linearity of detection
The use of RT-ddPCR is superior to qRT-PCR because of its quantifiability, and its tolerance of mismatched probe/primer sequences, as well as higher precision at low copy numbers, but without sacrificing sensitivity or reliability. In fact, the sensitivity was found to be up to 1-10 copies, with linearity over 3-4 orders of magnitude.
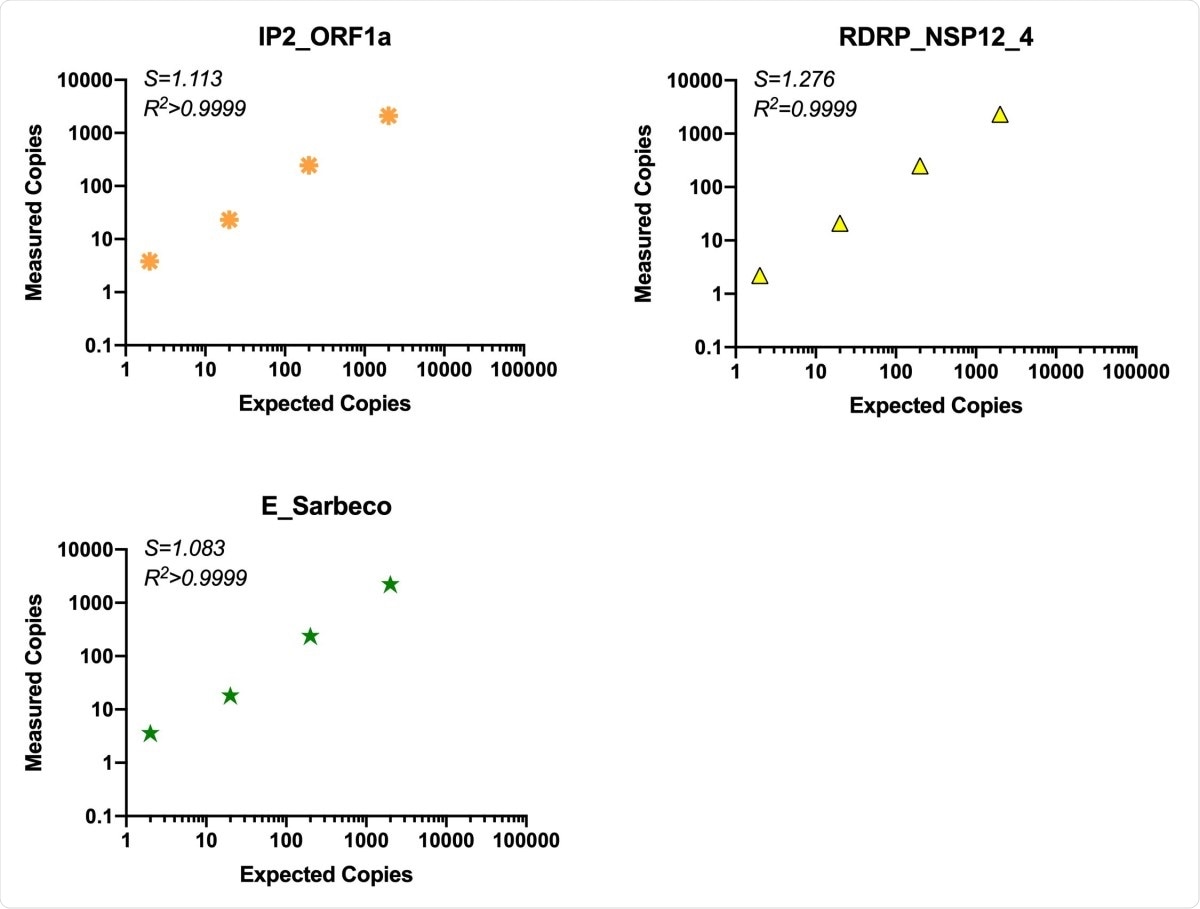
Comparison of assay efficiency and linearity of published assays, ORF1a “nCoV_IP2” and E gene and novel RDRP-NSP12 assay. The performance of our RDRP NSP12 assay was compared to published primers/probes for ORF1a and the E gene in the ddPCR platform using common RT reactions containing virion standard RNA inputs of 2-2x104 copies/ddPCR well. S (slope) and R2 are indicated for each assay.
Many assays have lost sensitivity as a result of mutations that affect primer annealing, which indicates the sensitivity of detection when multiple genomic regions are detected simultaneously, as with the current panel. High sensitivity, while not essential for the detection of infection, is likely to be very useful in revealing the course of the disease. The assays in current use are often found to become negative at 2-3 weeks following infection, or to alternate between positive and negative at successive time points. Some infected individuals continue to have prolonged viral shedding. The multi-target assay can also reveal the transcriptional profile of the virus.
As such, this panel of assays can be applied to a diverse range of clinically relevant samples in which SARS-CoV-2 RNA may be in low or high abundance.”
Higher copy numbers at 3’ UTR
The genomic regions at the 3’ UTR often had higher copy numbers compared to the 5’ UTR, perhaps because of increased efficiency of reverse transcription when assays are carried out at this end, due perhaps to the poly-dT sequence. Another explanation is that at this end, they detect sgRNAs within infected cells that have not yet been packaged into viral particles. These could have entered the supernatant from dying cells, or may be within the exosomes at low levels. Thus, cell-rich samples, or those with more cell-associated RNA, may have more sgRNA targets, which again may affect clinical tests and research results.
Background RNA enhances efficiency
The current assays are ideal for testing samples from a variety of tissues with significantly different gRNA levels, because, as with other studies, here too the researchers found that the tests worked best when some background RNA was present. This remained true even though assay efficiency was reduced as the RNA concentration went above 100ng/µL.
High comparability between RT-ddPCR and qRT-PCR
The results obtained by RT-ddPCR and qRT-PCR both show a strong correlation, while the comparison also confirms the ability of the former to yield trustworthy data even at low copy number.
Implications
The panel thus contains sensitive, quantitative assays for SARS-CoV-2 RNA that can be used to target the whole genome, detecting both genes and intergenic regions, structural and non-structural proteins, and genes in sgRNAs. The implications of sgRNA transcription are yet to be known. The current test panel may help understand how SARS-CoV-2 determines gene expression via varying levels of gRNAs and sgRNAs, and how this, in turn, influences the course of the disease.
These assays can serve as novel molecular tools to investigate SARS-CoV-2 infection, replication dynamics, and gene expression to better understand the viral dynamics and pathogenesis of SARS-CoV-2 over the course of infection.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Telwatte, S. et al. (2021). Novel RT-ddPCR assays for determining the transcriptional profile of SARS-CoV-2. bioRxiv preprint. doi: https://doi.org/10.1101/2021.01.12.425991, https://www.biorxiv.org/content/10.1101/2021.01.12.425991v1
- Peer reviewed and published scientific report.
Telwatte, Sushama, Holly Anne Martin, Ryan Marczak, Parinaz Fozouni, Albert Vallejo-Gracia, G. Renuka Kumar, Victoria Murray, et al. 2022. “Novel RT-DdPCR Assays for Measuring the Levels of Subgenomic and Genomic SARS-CoV-2 Transcripts.” Methods 201 (May): 15–25. https://doi.org/10.1016/j.ymeth.2021.04.011. https://www.sciencedirect.com/science/article/pii/S1046202321001031.