To date, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of coronavirus disease 2019 (COVID-19), has caused more than 93.21 million infections worldwide. As it spreads through different populations, it undergoes adaptations that sometimes affect its transmissibility and other biological characteristics.
Significance of adaptation studies
An important step in understanding any pandemic caused by a novel pathogen is to identify the changes that occur in the organism’s genome over time, and how these evolutions reflect changes in its behavior. This helps to pick out targets for intervention. The intense pace of research on the ongoing COVID-19 pandemic has led to the aggregation of thousands of complete viral RNA sequences, from multiple populations and regions.
The current study aims to exploit this database of viral genomic information to identify, first, the occurrence of genomic change in the SARS-CoV-2 as a result of global selection pressure, or regional selection, with alterations over time. Selection pressures are agents acting on the virus from outside, that affect its survival fitness positively or negatively by impairing or enhancing particular traits. The study of viral populations selected over a period of time could help uncover changes in virulence or immunogenicity associated with such genetic adaptive processes.
Phylogenetic approaches using deep learning
The researchers relied on phylogenetic methods to make their inferences, without using recombination data. Their approach is different from more conventional methods that summarize sequence data into numerical or graphical form, in order to identify how nucleotide variants are distributed in situations without selection pressure. In such neutrally evolving situations, free recombination is assumed to occur, with the population remaining constant.
In the current study, the researchers opted for phylogenetic techniques that require numerous repetitive events in a specific time interval in order to capture excess events beyond the limits of neutral evolution. This means that they are often used only with genomes that show a high rate of mutation, or have long trails of mutation.
The researchers used the ability of deep learning methods to capture complex genetic changes in prediction tools based on simulations, so that they do not have to spell out clearly defined parameters. Already, deep learning has been applied in population genetics to obtain predictions of various genetic parameters, such as recombination rate and selection, as well as germline data analysis. They can use image-recognition parameters to analyze viral adaptation, thus using the information in haplotype alignments.
Image-based haplotype analysis
The study is based on a convolutional neural network (CNN) combined with a recurrent neural network (RNN) approach. Called image-based haplotype guided evolutionary inference (ImHapE), this platform allowed them to identify selection in a quantitative manner in expanding viral populations, using genomic sequencing data. This four-step approach was modified to increase its speed while conserving its ability to capture differences in fitness in different populations in which selection is operating at different strengths. They simulated exponential growth in the population, with the rate of growth in positively selected populations being higher as fitness increased. Fitness is defined as “a reduction in death rate such that a fitness (1 + s) of 2 was equivalent to a 50% reduction in the viral death rate in the beneficial virus population.”
Once their CNN/RNN model had been trained and validated on their simulated population, they applied it to two sets of actual global data on the virus. The first was from the Global Initiative on Sharing All Influenza Data (GISAID) database, gathered between March and July 2020. The second was COG UK data, collected between April and December 2020. Mutations in the two databases were called using the Wuhan reference genome and an England reference genome, respectively.
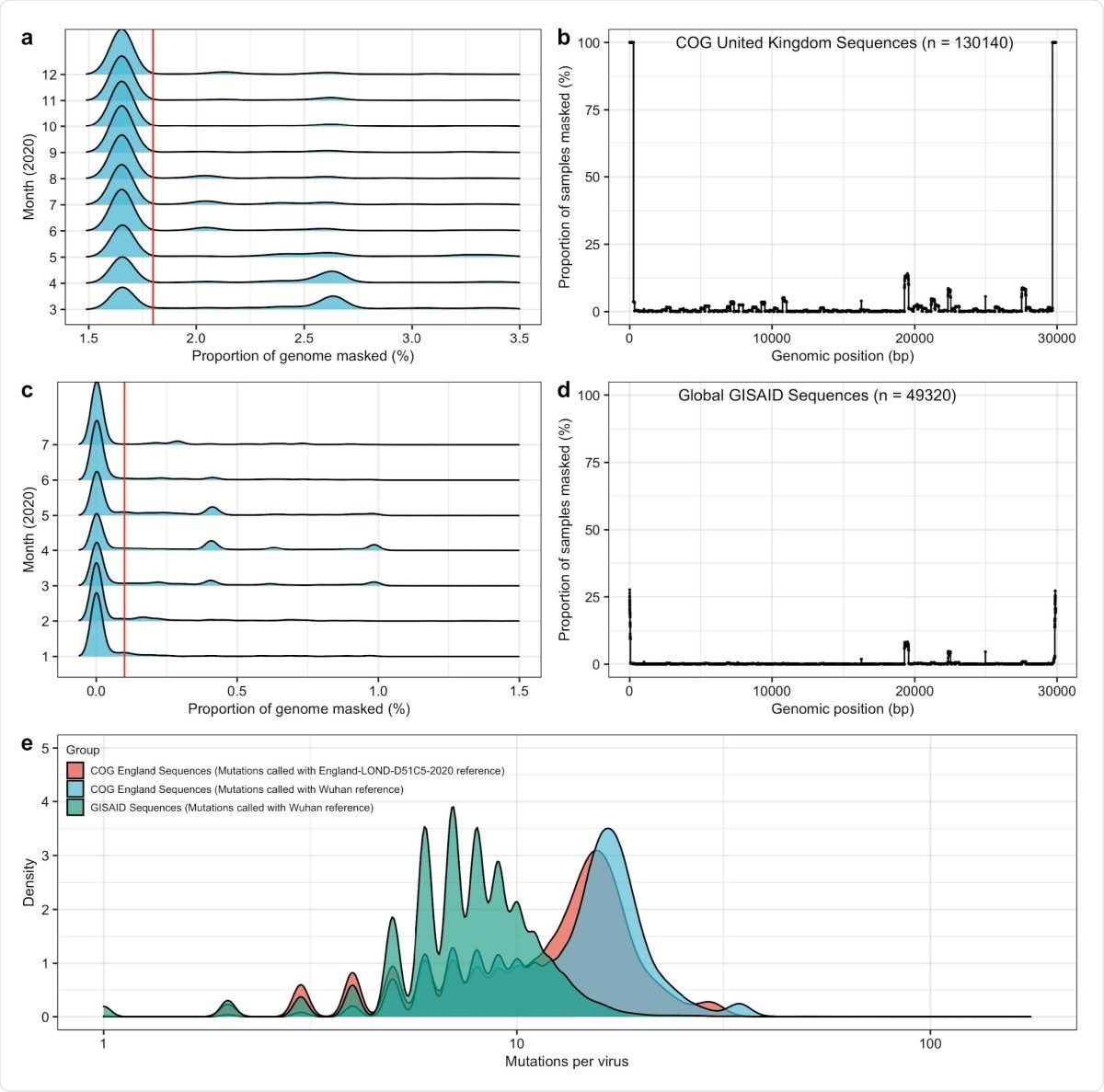
General quality control and sample filtering for GISAID and COG UK data. To ensure accurate inferences were being made in empirical data, we set a threshold for sample inclusion based on the proportion of genome masked. To set the threshold for the proportion of genome masked, we set simple cut-offs based on the entire sample distribution. For COG UK data, a, we only accepted samples with less than 1.8% of the genome masked, and b, did not find an excessive number of sites with masking in a genome-wide scan. For GISAID data, c, we only accepted samples with less than 0.1% of the genome masked, and b, did not find an excessive number of sites with masking in a genome-wide scan. e, We compared mutational counts in GISAID data (from January to August 2020) and COG UK data (from March to December 2020). In particular, we ensured that mutation counts using the LONGD51C5 reference genome from April 2020 resulted in a smaller number of mutations on average then mutations called n COG UK data using the Wuhan reference from December 2019.
They adjusted their mutation rate to fit with the estimated 23 mutations per genome per year that the SARS-CoV-2 genome is estimated to undergo. They also pooled virus samples according to region and time points.
Increased fitness of new variants
They found that the virus was undergoing positive selection in every population, as shown by a value over 1, but its fitness was different in different regions. Positive selection was observed even after compensating for population growth.
In Europe and North America, the fitness decreased over time, subsequent to the fixation of the D614G variant, but was higher in July than at the beginning. The fitness in March and July were 1.05 vs 1.42 in Europe, and 1.27 vs 1.40 in N. America.
Using the UK COG data, they found that positive selection had a large range of variation early in the outbreak. However, fitness increased from 1.05 at week 29 to 1.34 at week 49. The beginning of increased fitness was associated with the simultaneous expansion of the B1.177 lineage at week 29, with the new variant B.1.1.7 expanding after week 46.
The B. 1.177 lineage is defined by an A222V mutation in the spike protein. It has become widespread in Europe, but it is unknown whether this is related to any phenotypic advantage, such as increased transmissibility. The new variant, B.1.1.1.7, seems to be associated with increased fitness following week 46, indicating that this is fitter than the other circulating lineages in the UK at present.
Both the continental and the COG UK data profiling is an excellent example of how the simulation-based CNN/RNN tools can track selective differences among viral clones almost in real time.”
What are the implications?
These findings show how this tool is useful in following viral populations undergoing selection pressure, and thus making inferences about the selection-driven differences in virulence and infectivity. This versatile tool is ready to be refined and developed further to study adaptation in genomes.
Our general framework can be adapted and applied to any non-recombining population where aligned haplotype information is available such as somatic tissues or cancers.”

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.