Previous studies have demonstrated that patients with COVID-19 swiftly seroconvert to SARS-CoV-2, which means they produce Immunoglobulin IgA, IgG, and IgM antibodies directed against several viral proteins. Nonetheless, it is not completely clear whether all antibody responses are beneficial for us, or some can actually lead to a less favorable course of the disease.
Furthermore, enhancement of infection by antibodies has been reported in the medical literature for severe acute respiratory syndrome coronavirus (SARS-CoV), which is a causative agent of the original SARS epidemic and closely related to SARS-CoV-2.
Needless to say, that refinement of diagnostic, therapeutic and vaccination approaches against COVID-19 would benefit immensely from a clear and comprehensive understanding of all the correlates of immune response to SARS-CoV-2 infection.
Consequently, experts from the Antigen Discovery Incorporated, the University of California, CDC and Mayo Clinic in the US created and utilized a multi-coronavirus protein microarray that contains over one thousand full coronavirus proteins, protein fragments and peptides, in order to map IgM, IgG and IgA antibody epitopes COVID-19 patients' sera.
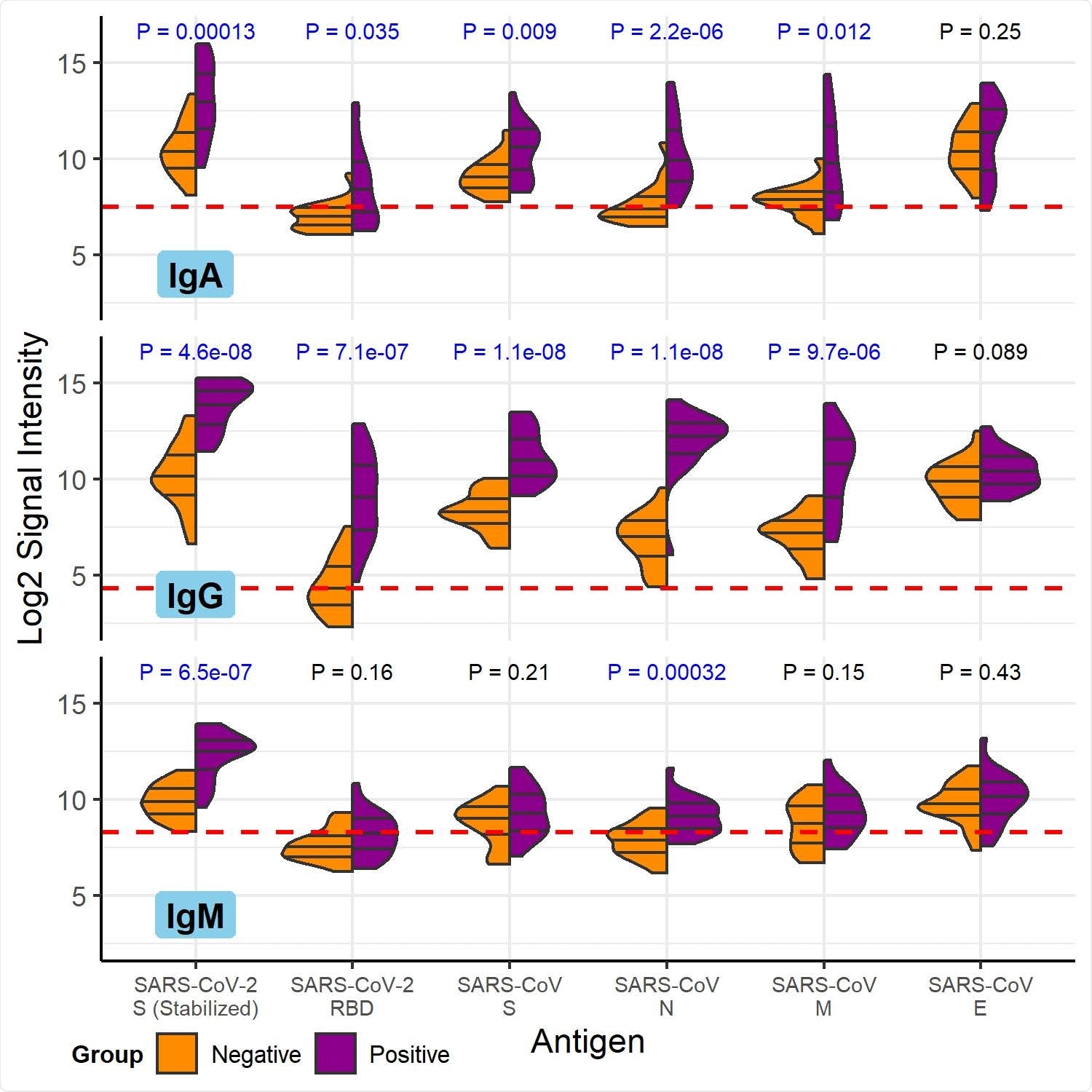
COVID-19 patient and healthy control antibody reactivity with purified SARS-CoV- 2 and SARS-CoV proteins. The split violin plot shows the log2-transformed fluorescence signal intensity distribution of antibodies bound to each purified protein on the multi-coronavirus protein microarray. Within each half-violin are three lines representing the interquartile range and the median. Above each split violin is the Wilcoxon rank sum p value, colored blue for significant p values below 0.05. The three panels are split by isotype (IgG, top; IgA, middle; IgM, bottom). Horizontal red dashed lines are drawn at the median of all signal intensities against purified proteins (n=14) and peptides (n=587) plus 1.0, i.e. double the global median—this threshold serves as a point of reference, but not necessarily a seropositivity cutoff for each protein.
Bacteria and cell lines as protein producers
The study approach localized the antibody reactivity of COVID-19 patients within SARS-CoV-2 proteins and allowed the researchers to map bound antigenic regions. Moreover, the study group was able to measure antibody reactivity of both COVID-19 patients and healthy individuals with seasonal human coronaviruses and with the two previous epidemic coronaviruses.
All coronavirus proteins that were used in this study were produced in the bacterial species Escherichia coli – except the SARS-CoV and SARS-CoV-2 spike glycoproteins, which were made in Sf9 insect cells, and the SARS-CoV-2 receptor binding domain, which was made in human embryonic kidney 293 cells.
Furthermore, most samples used in this study were obtained from patients who were not hospitalized, with blood collected between 26 and 60 days after symptom onset. Negative controls were collected before the COVID-19 pandemic, i.e., in the fall of 2019.
Validated enzyme-linked immunosorbent assay (ELISA) was used to test samples against the pre-fusion stabilized ectodomain of SARS-CoV-2 spike protein, and SARS-CoV-2 microneutralization assays was also pursued following biosafety level-3 precautions.
The correlates of cross-reactivity
After studying twenty COVID-19 patients, the researchers found the most robust antibody responses steered towards the SARS-CoV-2 proteins directed to the nucleocapsid (N) and S2 proteins for all antibody isotypes, akin to other studies in the field.
Antibody responses to M, S1, and accessory proteins 3a and 7a were also detected. Likewise, the scientists were able to localize regions of each of these SARS-CoV-2 proteins to which antibodies bound by analyzing antibody reactivity with overlapping protein fragments of three different lengths: 100, 50, and 30 amino acids.
"Our results were internally consistent in that reactive proteins had more reactive fragments than non-reactive proteins and 100 amino acid reactive fragments contained reactive 50 amino acid fragments and sometimes they also contained reactive 30 amino acid fragments", say study authors.
Manifold proteins of SARS-CoV, MERS-CoV and the seasonal coronaviruses HCoV-NL63 and HCoV-OC43 were also more reactive with IgM, IgG and IgA in COVID-19 patient sera when compared to the unexposed control sera, lending further credence to potential immunologic cross-reactivity between these viruses.
Useful biomarkers of infection and protection
In short, this type of multi-coronavirus protein array represents a tool that can help the scientific community improve the understanding of the immune response to SARS-CoV-2 and other coronaviruses.
"With these first two sets of convalescent sera provided by the Mayo Clinic and the CDC, we have shown that SARS-CoV-2 naïve subjects have clearly measurable cross-reactive antibodies to the whole N and S2 proteins and that this reactivity is limited to specific epitopes", explain researchers in this medRxiv paper.
What is important is that there are epitopes more specific to SARS-CoV-2 that might be viewed as useful biomarkers of infection. On the other hand, infection with SARS-CoV-2 can give rise to antibodies that bind to the nucleocapsid and S2 proteins of other coronaviruses – including HCoV-NL63, HCoV-OC43, SARS-CoV and MERS-CoV.
Finally, if high antibody levels developed to specific epitopes are found to be particularly protective, further research is needed to enable the use of this array for screening convalescent plasma with therapeutic potential, as well as vaccine recipient sera as an early efficacy measure.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Camerini, D. et al. (2020). Mapping SARS-CoV-2 Antibody Epitopes in COVID-19 Patients with a Multi-Coronavirus Protein Microarray. medRxiv. https://doi.org/10.1101/2021.01.14.21249690, https://www.medrxiv.org/content/10.1101/2021.01.14.21249690v1
- Peer reviewed and published scientific report.
Camerini, David, Arlo Z. Randall, Krista Trappl-Kimmons, Amit Oberai, Christopher Hung, Joshua Edgar, Adam Shandling, et al. 2021. “Mapping SARS-CoV-2 Antibody Epitopes in COVID-19 Patients with a Multi-Coronavirus Protein Microarray.” Edited by Alison Sinclair. Microbiology Spectrum 9 (2). https://doi.org/10.1128/spectrum.01416-21. https://journals.asm.org/doi/10.1128/Spectrum.01416-21.