To date, the coronavirus disease 2019 (COVID-19) pandemic has spread to 191 countries and infected more than 95.62 million people. Of these, more than 2 million people have lost their lives. The United States remains the nation with the highest number of cases, reaching 24 million, while India and Brazil follow, with over 10.58 million and 8.51 million cases, respectively.
New strains of the virus have begun to emerge with increased infectivity – including the VUI 202012/01 in Britain and 501.V2 in South Africa. This has led to concerns among public health authorities that mutant strains may evade neutralizing antibodies produced by convalescent individuals as well as the present vaccine candidates.
Many countries, including Britain and South Africa, are experiencing a successive wave of COVID-19 cases, with skyrocketing numbers being reported in many parts of the world.
To better account for these risks, US-based researchers at the Fred Hutchinson Cancer Research Center, the University of Washington, Vanderbilt University, and Harvard Medical School, have systematically identified potential SARS-CoV-2 mutations that could evade neutralizing antibody detection.
The study
SARS-CoV-2 acquires mutations that can prevent the antibody response. However, scientists can predict what mutations may emerge as the virus evolves.
The researchers aimed to map how the 4,000 potential mutations to the SARS-CoV-2 spike receptor-binding domain (RBD) can impact antibody binding and neutralization.
To arrive at the study findings, which were published in the journal Cell Host Microbe, the researchers developed a deep mutational scanning method to map how all amino-acid mutations in the RBD impact antibody binding. They also wanted to apply the method to ten human monoclonal antibodies.
To do this, they expressed the mutant variants of RBD on the surface of yeast cells and exposed them to ten antibodies, which were extracted from COVID-19-confirmed patients. The team captured the cells that manifested impaired binding to the antibodies and conducted deep sequencing to determine the RBD mutations existing in these cells.
“The complete escape maps predict which mutations are selected during viral growth in the presence of single antibodies,” the researchers noted in the paper.
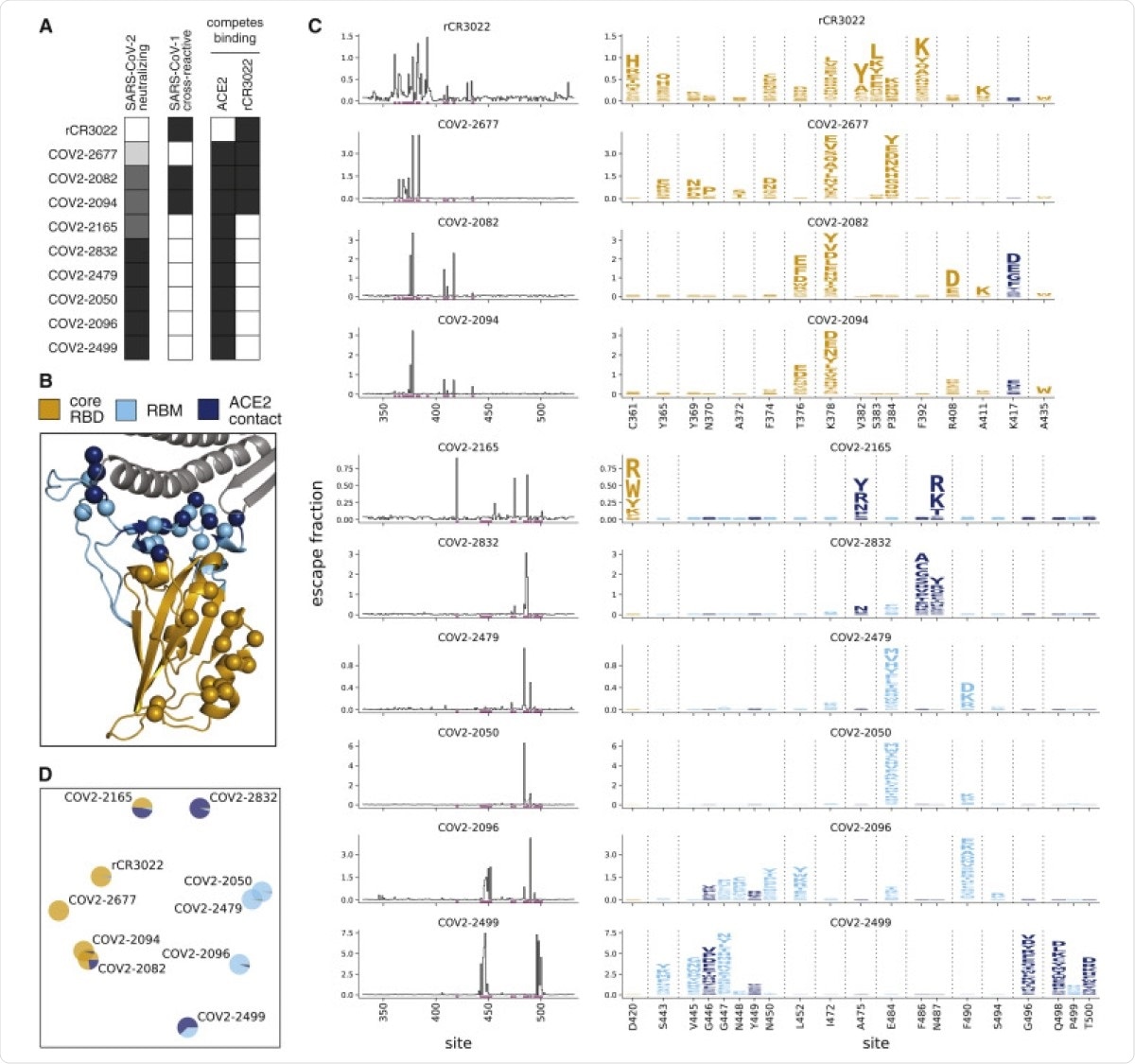
Complete Maps of Escape Mutations from 10 Human Monoclonal Antibodies. (A) Properties of the antibodies as reported by Zost et al. (2020a). SARS-CoV-2 neutralization potency is represented as a gradient from black (most potent) to white (non-neutralizing). Antibodies that bind SARS-CoV-1 spike or compete with RBD binding to ACE2 or rCR3022 are indicated in black. (B) Structure of the SARS-CoV-2 RBD (PDB: 6M0J; Lan et al., 2020), with residues colored by whether they are in the core RBD distal from ACE2 (orange), in the receptor-binding motif (RBM, light blue), or in direct contact with ACE2 (dark blue). ACE2 is in gray. RBD sites where mutations escape antibodies are indicated with spheres. (C) Maps of escape mutations from each antibody. The line plots show the total escape at each RBD site (sum of escape fractions of all mutations at that site). Sites with strong escape mutations (indicated by purple at bottom of the line plots) are shown in the logo plots. Logo plots are colored by RBD region as in (B). Different sites are shown for the rCR3022-competing antibodies (top four) and all other antibodies (bottom six). For interactive escape maps, see https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_Crowe_antibodies. (D) Multidimensional scaling projection of the escape mutant maps, with antibodies having similar escape mutations drawn close together. Each antibody is shown with a pie chart that uses the color scale in (B) to indicate the RBD regions where it selects escape mutations. See also Figures S1 and S2 and Table S1.
“They further enable the design of escape-resistant antibody cocktails-including cocktails of antibodies that compete for binding to the same RBD surface but have different escape mutations,” they added.
Aside from that, the team also identified some locations on the protein where these escape mutations are grouped. They observed that the mutations tended to differ between various antibodies.
The team concluded that the complete escape-mutation maps can help develop new therapies and vaccines that can adapt to viral evolution. The researchers also said that they are working on expanding the new approach, to address the largest problem on SARS-CoV-2 evolution.
Our next steps will be to apply this approach to serum from individuals who have been vaccinated against SARS-CoV-2 to see how mutations might reduce binding and neutralization by vaccine-elicited antibodies,” Allison Greaney, a study co-author, said.
Source:
Journal reference: