The main thrust of scientific research on therapeutics to counter the ongoing coronavirus disease 2019 (COVID-19) pandemic has been aimed at identifying drugs and antibodies that can act against the entry of the causative virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), into the host cell.
A new study by US-based researchers shows the presence of protective antibodies that target the N-terminal domain (NTD) of the spike instead of the spike receptor-binding domain (RBD).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The team – from Vanderbilt Vaccine Center, the University of Washington School of Medicine, St. Louis, and Integral Molecular – have published their findings on the preprint bioRxiv* server.
Spike protein
Viral entry is mediated by the spike protein of the virus, which binds to the host cell receptor, angiotensin converting enzyme 2 (ACE2), via the RBD. The spike is a trimeric glycoprotein with two subunits to each protomer, the S1 and the S2. The S1 contains the RBD, and is responsible for binding the ACE2.
The S2 subunit mediates virus-cell membrane fusion and viral entry into the cell. Most neutralizing antibodies have therefore targeted the spike RBD.
The RNA genome and protein transcriptome of the virus has changed thousands of times, with an average of four mutations per amino acid in the spike protein. This has aroused concern as to whether viral variants would allow escape from neutralizing antibodies, most of which bind to the RBD. Some scientists have reported monoclonal antibodies (mAbs) that bind to regions outside the RBD, too.
Neutralizing activity
The current study identifies the structure and functionalities of three anti-NTD antibodies that were selected from a library of around 390 mAbs obtained from individuals who had recovered from natural SARS-CoV-2 infection. Two of the three had powerful neutralizing capacity. These were named COV2-2676 and COV2-2489.
They found that in two types of neutralization assays, a focus reduction neutralization test (FRNT) using the live virus, and the second a real-time cell analysis (RTCA) assay that used the spike-expressing recombinant replication-competent vesicular stomatitis virus (VSV) pseudovirus. They found that both the mAbs were able to neutralize the wildtype virus in the FRNT in a dose-dependent manner, with the half-maximal inhibitory (IC50) value being 501 or 199 ng/mL, respectively. In the second, both neutralized the pseudovirus with IC50 being 8 or 56 ng/mL, respectively.
Common binding region
The scientists identified the epitopes that bound these potent neutralizing antibodies. Both bound to a distinct antigenic site compared to the epitopes bound by anti-RBD human mAbs. Both these antibodies failed to prevent soluble ACE2-soluble RBD binding. In fact, both antibodies attached to the NTD of the spike in the ‘closed’ conformation of the spike trimer. They compete fiercely with each other because of the common binding epitope. Both are independent clonotypes, encoded by the heavy chain variable gene segment IGHV-4-39 and IGHV1-69, respectively. Their HCDR3 regions are quite different, however.
Recognition of SARS-CoV-2 spike NTD
The heavy loops of both these antibodies interact with the spike NTD at the N3 and N5 loops, respectively, showing that this is a potential therapeutic target, and that individuals with acquired immunity to the virus produce antibodies targeting this region.
Using alanine scans, the researchers also determined that single alanine changes at several positions involved in the binding of these mAbs did not impair the binding of another anti-NTD antibody, COV2-2305, probably because these loops contain the key contact residues at strategic locations. Using an RTCA assay, they found escape mutations that resisted neutralization by these antibodies at levels of 10 or 100 μg/mL of COV2-2676 and COV2-2489, respectively. The escape mutations were in the same region identified by the alanine scan. Thus, both methods indicate a common antigenic site of binding for both antibodies, with different epitopes within this region.
Binding of spike to infected cells
They found that these mAbs bound avidly to spike proteins on the surface of infected cells, with greater efficiency and density of binding compared to antibodies that targeted the RBD. They repeated the FRNT on several cell lines cultured with the virus. They found, however, that this binding varies with cell type, perhaps because the entry factors and receptors for the virus differ between different kinds of cells.
They also compared the results of incubating the mAbs with the virus before and after it was absorbed on the cell surface. They found that the use of these antibodies produced potent neutralization both pre- and post-attachment to the virus. These findings suggest that they could be used to prevent infection even after the virus attaches to the cell.
Neutralization was not observed with these mAbs in cells that overexpressed human ACE2 and TMPRSS2, though anti-RBD antibodies inhibited virus attachment.
Full-length or F(ab’)2 forms required
Both these mAbs were able to neutralize the virus only in the full-length IgG or divalent Fab form, but the monovalent Fab fragments of either were unable to neutralize it. In contrast, Fabs of potently neutralizing anti-RBD antibodies inhibited infection as efficiently as the full-length antibody.
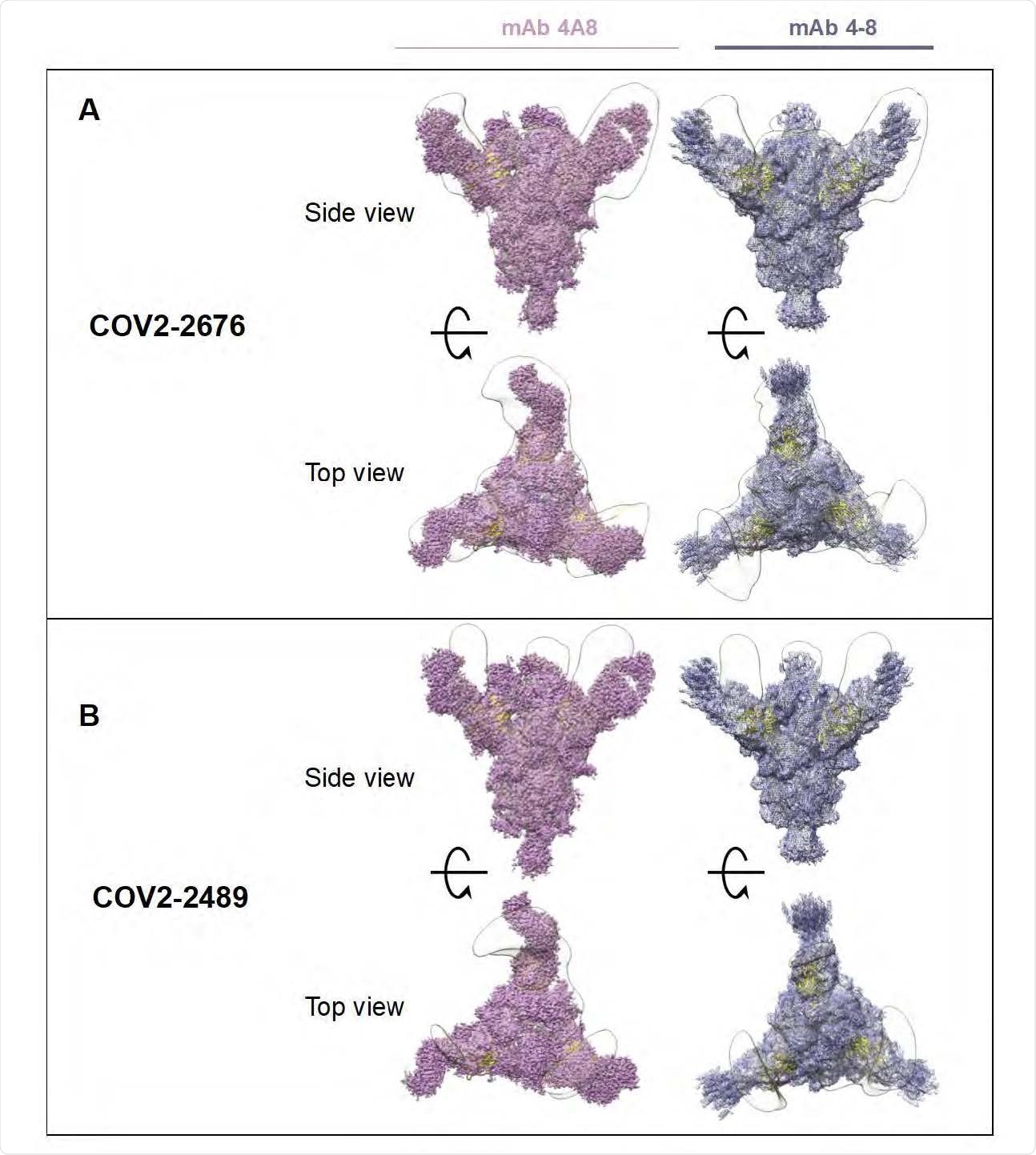
Superimposed Fab-Spike negative stain EM. A) COV2-2676 with mAb on the top is side view 4A8 (left), mAb 4-8 (right) and bottom is top view of the same. B) COV2-2489 with mAb on the top is side view 4A8 (left), mAb 4-8 (right) and bottom is top view of the same.
The researchers hypothesize, “The larger IgG or F(ab’)2 might sterically hinder functional interactions of the S protein in entry. Alternatively, the monovalent Fab molecule may bind with too low an affinity to the NTD to inhibit entry.”
Fc-mediated mechanism
These antibodies required intact Fc-mediated functions for their activity in vivo. When these interactions were abolished, the mice developed greater weight loss, viral burden was higher, and lung inflammation was higher.
Protective effect in mice
The researchers found that these mAbs protected mice expressing the human ACE2 receptor when given before or after a viral challenge. Treated mice did not show weight loss, reduced the viral burden, and reduced cytokine levels, similar to the effects of an anti-RBD antibody.
What are the implications?
Regions of the NTD are recognized by neutralizing and protective antibodies against SARS-CoV-2 and could function as part of antibody cocktails to minimize the selection of escape variants or resistance to natural variants in RBD as they emerge.”
The researchers recommend the use of a combination of antibodies targeting the spike RBD and the NTD to boost the level of protection, reduce the risk of emergence of escape mutations, despite the relatively lower potency of neutralization offered by the anti-NTD antibodies compared to the most potent neutralizing anti-RBD antibodies. Vaccines may also offer the most durable immunity if they are developed against multiple antigens, rather than only the spike RBD.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources