The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes the coronavirus disease 2019 (COVID-19), is a novel pathogen that emerged in December 2019 in China. Since then, it has spread globally to over 191 countries and territories, infecting more than 96.82 million people and claiming over 2 million lives.
As the virus continues to spread and new highly infectious variants emerge, understanding the complex pathogen-host interplay is urgently needed to effectively control its spread. Pathogens, such as viruses and bacteria, assume host pathways to trigger a tolerant environment for their proliferation.
Researchers at the Department of Ophthalmology and Visual Sciences, the University of Illinois at Chicago, USA, provided an overview of SARS-CoV-2 and its life cycle pathways. In doing so, the team addressed the functional changes in the viral proteins, providing important strategic clues to help that may help in the ongoing development of therapies against COVID-19.
SARS-CoV-2 proteins
Both the severe acute respiratory syndrome coronavirus (SARS-CoV) and the SARS-CoV-2 belong to the beta subfamily of the coronavirus genus. SARS-CoV-2 contains 29 different proteins, which include 16 nonstructural proteins. These include proteases, RNA-dependent RNA polymerases, a nuclease, helicase, and methyltransferase encoded by 14 open reading frames (ORFs).
SARS-CoV-2 has the highest transmission rate among the coronaviruses that have caused outbreaks in the past. However, it has the lowest mortality rate of about 2.3 percent, compared to SARS-CoV or the Middle East respiratory syndrome coronavirus (MERS-CoV), with a fatality rate of 9.6 percent and 35 percent, respectively.
The reason why SARS-CoV-2 has the highest rate of transmissibility is that it has genetic alterations in the various structural and nonstructural proteins.
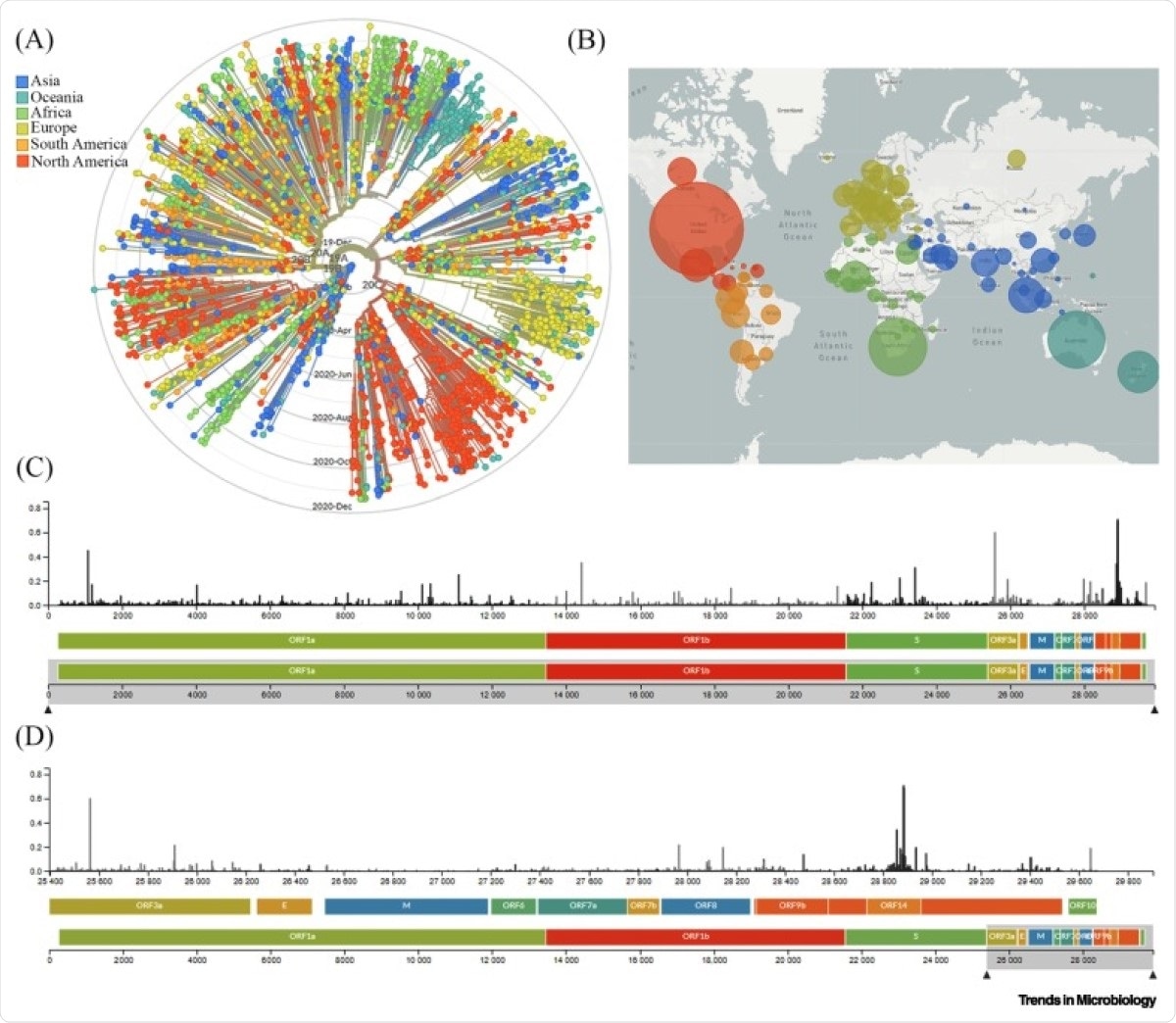
Global Genomic Epidemiology of Novel Coronavirus Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2). (A) Phylogenetic analysis of 3485 genomes of SARS-CoV-2 sequences globally from December 2019 to December 2020. The different genetic variants of circulating SARS-CoV-2 are grouped into five clades defined by specific signature mutations showing their global distribution on the time scale. The clades 19A and 19B dominated the early outbreak in Wuhan and represent a higher proportion in Asia. Clades 20A, 20B, and 20C dominate in Europe and North America. (B) Geographical distribution of genomes. Each circle is centered on an individual country. The color indicates the region, and the size (area) of the circle represents the number of genomes from that country. (C) A ‘diversity’ panel that shows the novel coronavirus genome, its genes, and sites of amino acid mutations. (D) Subsection of subfigure (C) highlighting the mutation pattern in the 25 400–29 800 bp range of the genome. Apart from the spike (S) region, the genomic regions of open reading frame (ORF)14, ORF9b, ORF8, and ORF3a appear to be highly variable between clinical isolates of SARS-CoV-2. Source: latest global SARS-CoV-2 updated daily at https://nextstrain.org/sars-cov-2. Abbreviations: E, envelope; M, membrane.
The study
In the study, which was published in the journal Trends in Microbiology, the team highlights the importance of understanding the mechanisms by which the virus triggers host cell pathways and functions to cause illness.
The team discussed genomic variations in viral proteins to determine their roles in the modulation of host cell signaling.
Among the key findings of the study was that SARS-CoV-2 proteins modulate host signaling to produce a permissive or tolerant environment. This way, the environment in the host can support viral proliferation and spread.
Further, the team found that viral proteins can boost or lower the activity of about 100 human kinases involved in cellular function, metabolism, and immune activation. This means that the proteins of SARS-CoV-2 alter the body and cellular functions to aid in its spread.
The team also noted that targeting particular proviral cellular signaling could help make the host environment resistant to virus replication. Therapeutics developed for this function can aid in containing virus spread and preventing infection.
The nonstructural proteins of SARS-CoV-2 also participate in dysregulating the immune response. This leads to a weaker activity of the immune system in fighting the infection. The virus alters innate cellular defenses using many of its proteins, leading to a delayed hyper inflammation along with a weakened interferon (IFN) response.
When immune activation is delayed, it prolongs infection and promotes viral replication. A substantial IFN production may ensue, leading to a cytokine storm that may lead to acute tissue injury.
Collectively, it is clear that individual factors of SARS coronaviruses have diverse roles in disruption of cellular programs, and additional research focus directed in the areas outlined here may help in delineating the underlying mechanism of pathogenesis and spread in humans and will be instrumental in our quest for curative therapeutics,” the researchers concluded in the study.
Understanding how SARS-CoV-2 affects the body can help in the development of vaccines and therapies to fight the ongoing pandemic.
Source:
Journal reference: