The etiological agent of coronavirus disease 2019 (COVID-19), the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), transmits efficiently from human-to-human. Since the virus was first detected in December 2019 in Wuhan, China, new variants – with mutations at the virus’s spike protein – have emerged, resulting in heightened transmissibility. With no targeted and effective antiviral treatment options currently available, mass vaccinations remain our best bet at turning the tide of the COVID-19 pandemic.
While nations worldwide are partaking in the enormous COVID-19 vaccination plans, perfecting the rapid roll-out of an efficacious vaccine in the face of numerous infectious diseases calls for further research and development in the field.
Researchers in Singapore have designed a subunit vaccine based on the SARS-CoV-2 spike protein co-administered with CpG (Oligodeoxyribonucleotides) adjuvant. The researchers encapsulated both antigen and adjuvant with their proprietary artificial cell membrane (ACM) polymersome technology. This encapsulation enhances the immunogenicity of their formulation. The team has reported their promising findings in a recent bioRxiv* preprint paper.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
They found that their formulation elicited robust neutralization against SARS-CoV-2 in C57BL/6 mice. This requires two doses of the prepared vaccine and the effect persisted for at least 40 days. Furthermore, they also confirm the presence of memory CD4+ and CD8+ T cells that produce Th1 cytokines. The research team’s model demonstrates effective and durable humoral and cellular immunity against SARS-CoV-2.
The SARS-CoV-2 belongs to the genus Betacoronavirus within the family Coronaviridae. Each virion is a nucleocapsid protein encapsulating the single-stranded genomic RNA, surrounded by a lipid bilayer.
Into this lipid bilayer, the spike (S), membrane and envelope proteins are incorporated. The spike is a trimer protein with a receptor-binding domain (RBD) containing two subunits: S1 and S2. The spike protein’s RBD enables viral entry into the host cell by interacting with the angiotensin-converting enzyme 2 (ACE2) receptor expressed on host cells.
The host proteases cleave the spike protein at the S1-S2 junction and induce significant structural rearrangement that exposes the hydrophobic fusion peptide, thus permitting the merging of viral and host cell membranes leading to viral entry.
Because the spike protein is immunogenic and the target of antibodies and T cells – particularly CD4+ T cells – it has emerged as the key target for subunit vaccines of various modalities.
The researchers investigated the immunological effect of the artificial cell membrane (ACM) polymersomes on the different SARS-CoV-2 spike proteins (the ectodomain of the spike protein, the S2 domain only, and a trimeric spike protein).
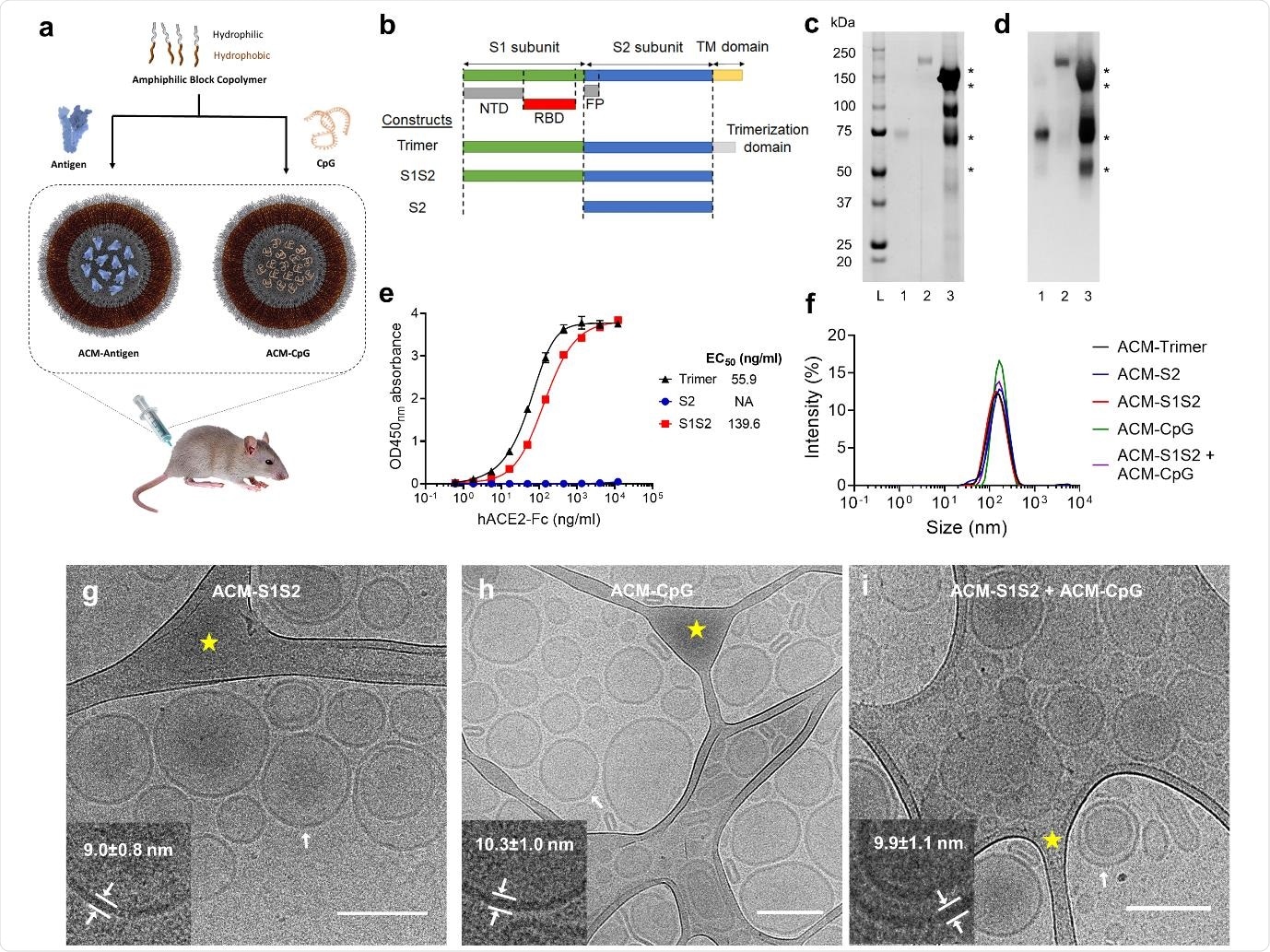
ACM-vaccine characterization. a. Schematic illustration of ACM-vaccine preparation. Antigens and CpG adjuvant were encapsulated within individual ACM polymersomes. A 50:50 v/v mixture of ACM-Antigen and ACM-CpG was administered to mice as the final vaccine formulation. b. Schematic of the spike protein variants used in this study. S1S2 protein was expressed and purified inhouse whereas S2 and trimer were purchased from commercial vendors. NTD: N-terminal domain. RBD: receptor binding domain. FP: fusion peptide. TM: transmembrane. c. SYPRO Ruby total protein stain. Lane L: Precision Plus Protein Standards (Bio-Rad). Lane 1: S2. Lane 2: trimer. Lane 3: S1S2. d. Western blot using mouse immune serum raised against SARS-CoV-2 spike. Western blot-reactive S1S2 bands are indicated by *. e. ACE2 binding curves of trimer, S2 and S1S2. f. Dynamic Light Scattering (DLS) measurements of ACM-antigens (ACM-trimer, ACM-S2 and ACM-S1S2), and ACM-CpG. ACM particles were determined to be 100-200 nm in diameter. g-i. Cryo-EM images of ACM-S1S2, ACM-CpG, and mixture of ACM-S1S2 + ACM-CpG illustrate the vesicular architecture with an average diameter of 158±25 nm (scale bar 200 nm). Inserts (lower left of each image) are magnifications of the bilayer membrane of vesicles at regions indicated by white arrows. Areas highlighted by yellow star are lacy carbon.
The researchers generated the spike protein, by engineering T.ni cells to express a spike variant that retained S1 and S2 domains. It excluded the hydrophobic transmembrane domain, thereby improving protein solubility. They used a commercial S2 fragment and a trimeric spike protein as controls. The S2 served as an ideal negative control since it lacked strongly neutralizing epitopes, whereas the trimeric spike was used as a positive control.
The traditional approaches of vaccine models – using inactivated or live attenuated virus – require biosafety level (BSL) 3 facility to handle SARS-CoV-2. The current mRNA vaccines require extremely cold conditions for maintenance to retain their stability. The high cost of the vaccine is also a setback. While the development of subunit vaccines is highly accelerated, some leading vaccine candidates are yet to overcome certain limitations.
Nanotechnology comes to the rescue: to develop a safe, cost-effective and scalable vaccine platform. Such a straightforward route, to well-defined nanoscale vesicles, is achieved with amphiphilic block copolymer self-assembly. Here, one can tune the constituent block concentration, membrane thickness and properties (including size and surface).
In this study, the researchers use their proprietary artificial cell membrane (ACM) – self-assembling nanoscale vesicles. These are polymersomes, structurally made up of an amphiphilic block copolymer that comprises polybutadiene-b-polyethylene glycol (PBD-PEO) and a cationic lipid 1,2- dioleoyl-3-trimethylammonium-propane (DOTAP). These serve as delivery vehicles that are efficiently taken up by dendritic cells, DC1 and DC2. Uptake up dendritic cells, the most efficient antigen-presenting cells (APC), are key to initiating the adaptive immune response.
The researchers previously established that the immunogenicity of a protein could be significantly improved through encapsulation within ACM polymersomes. The CpG used here is Murine CpG 1826.
Here, the researchers developed a subunit vaccine based on the spike protein of SARS-CoV-2, co-administering with CpG adjuvant. They have demonstrated the flexibility of the technology by encapsulating different classes of biomolecules (DNA and protein) within their proprietary ACM polymersomes to produce coherent and immunogenic particles.
The researchers are currently doing further titration experiments required to determine the optimum dose for a high-quality antigen. Based on the observations of this study, they propose this investigation on the use of ACM technology to address limited antigen availability in a pandemic.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Jian Hang Lam, Amit Kumar Khan, Thomas Andrew Cornell, Regine Josefine Dress, Teck Wan Chia, Wen Wang William Yeow, Nur Khairiah Mohd-Ismail, Shrinivas Venkatraman, Kim Tien Ng, Yee-Joo Tan, Danielle E. Anderson, Florent Ginhoux, Madhavan Nallani. Next-generation vaccine platform: polymersomes as stable nanocarriers for a highly immunogenic and durable SARS-CoV-2 spike protein subunit vaccine. bioRxiv 2021.01.24.427729; doi: https://doi.org/10.1101/2021.01.24.427729, https://www.biorxiv.org/content/10.1101/2021.01.24.427729v1
- Peer reviewed and published scientific report.
Lam, Jian Hang, Amit K. Khan, Thomas A. Cornell, Teck Wan Chia, Regine J. Dress, Wen Wang William Yeow, Nur Khairiah Mohd-Ismail, et al. 2021. “Polymersomes as Stable Nanocarriers for a Highly Immunogenic and Durable SARS-CoV-2 Spike Protein Subunit Vaccine.” ACS Nano 15 (10): 15754–70. https://doi.org/10.1021/acsnano.1c01243. https://pubs.acs.org/doi/10.1021/acsnano.1c01243.