Over a year since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the etiological agent of coronavirus disease 2019 (COVID-19) – the exact origins of this novel virus remains a mystery, with no clear animal progenitor or intermediary host confirmed.
A zoonotic (animal-derived) pathogen, SARS-CoV-2 was initially thought to have spilled over to human hosts through a “wet market” in Wuhan, China, where it was first detected in December 2019.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Environmental samples from the Huanan Seafood Market in the Jianghan District of Wuhan City showed evidence for infections in humans. However, subsequent case studies have found no links to this market. This suggests that it may not have been the “ground zero” for the SARS-CoV-2 spillover.
An early notion was that pangolins – widely sold and consumed in China – may have been the intermediary host that facilitated SARS-CoV-2’s jump to humans. This was hypothesized on the basis that pangolins are often reservoirs for coronaviruses. However, subsequent research has not corroborated this.
Moreover, the recent identification of SARS-CoV-2’s closest known relatives in horseshoe bat samples in Yunnan province, which is approximately 1,500 km from Wuhan, and as well as in Cambodia and Japan, suggest that these animal-derived pathogens have been circulating among bat species a lot longer than previously thought – across large geographical zones in China and beyond.
What studies have so far established, however, is that SARS-CoV-2 is a member of the Sarbecovirus subgenus of Betacoronaviruses seen in horseshoe bat hosts of the family Rhinophilidae, and a sister lineage of SARS-CoV, which caused the SARS outbreak in 2002-2003.
Now, a team of researchers in the UK and China performed a comprehensive analysis of horseshoe bat and pangolin Sarbecoviruses. The findings of their study have been published on the bioRxiv* preprint server.
Analyzing horseshoe bat and pangolin Sarbecoviruses
The researchers focused on a wider group of ‘nCoV’ Sarbecoviruses that cluster phylogenetically with SARS-CoV-2 and involved recombination detection analysis of the genome alignment of all available Sarbecoviruses. The team identified 16 recombination breakpoints that can help split the alignment into 17 putatively non-recombinant genomic regions from which the evolutionary history of the virus can be determined.
To characterize the recombination patterns between viruses of the same clade as SARS-CoV-2 and those in the same clade as SARS-CoV, the researchers classified each virus in each of the 17 regions to either belonging to the nCoV clade or the non-nCoV clade (which will be closer to SARS-CoV).
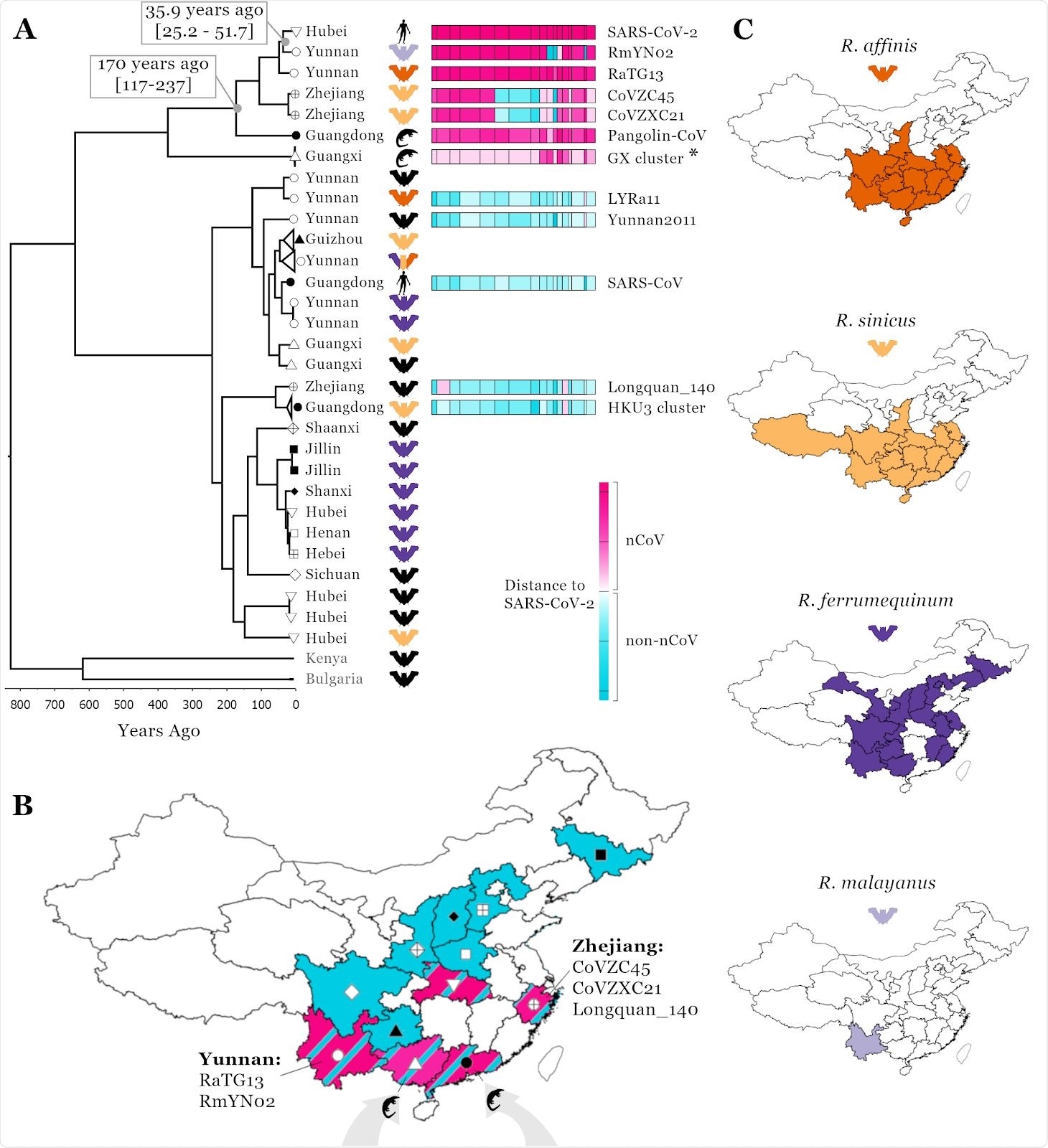
Linking SARS-CoV-2 related Sarbecovirus to geography and host ranges. Recombination analysis on a whole genome alignment of 69 Sarbecoviruses identified 16 recombination breakpoints (A), see methods. The fourth region was used for a molecular clock analysis (see scale below phylogenetic tree) and the median node age is presented for two key nodes including 95% HPD intervals; a representative set of 32 viruses are shown in the phylogenetic tree. T o illustrate recombination patterns between viruses in the same clade as SARS-CoV-2 and those in other clades of the tree, we have attributed each region of each virus to either being in the nCoV clade (the clade SARS-CoV-2 is found in, pink) or the non-nCoV clade (closer to SARS-CoV, blue). The colour shade corresponds to genetic distance, see key. The ‘GX cluster’ (asterisk) includes five viruses sampled in Sunda pangolins in Guangxi (P1E, P2V, P4L, P5E, P5L; see Table S1). A map of China (B) is shown with colours corresponding to regions Sarbecoviruses have been sampled: blue, the non-nCoV clade and pink, the nCoV clade. Symbols correspond to provinces at tree tips in A. Host ranges of four potential hosts of the proximal SARS-CoV-2 ancestor (C); ranges from Smith and Xie (2013).
Meat consumption led to ecological disturbances in SARS-CoV-2 transmission to humans
The work confirmed that Rhinolophus affinis is still the likely reservoir species as its range extends across Central and Southern China, which explains the bat Sarbecovirus recombinants in West and East China, bat Sarbecovirus recombinants linked to Southern China, and trafficked pangolin infections. As changes in meat consumption resulted in ecological disturbances, it led to SARS-CoV-2 transmission to humans through direct or indirect contact with the wildlife, and subsequent emergence in Hubei, Central China.
According to the authors, the only way of finding the animal progenitor of SARS-CoV-2 and the whereabouts of its close relatives is by increasing the sampling intensity. It is worth noting that the close relatives of SARS-CoV-2 may also have pandemic potential and pose a similar threat of emergence in animals and human population.
Pangolin’s susceptibility to an apparently new human coronavirus is not surprising given the well-documented generalist nature of SARS-CoV-2, which has been found to readily transmit to multiple mammals with similar ACE2 receptors and poses a grave risk of reverse-zoonosis as has been seen most notably with human to mink transmissions.”
The need for increased sampling
The data currently available shows a complex history behind SARS-CoV-2’s natural evolution, driven by co-circulation of related coronaviruses over the last century in bat populations across East, West, Central and Southern China. Multiple recombination events evident in the genomes of viruses sampled from different bat hosts in various parts of China indicate constant movement of these viruses due to the different bat populations carrying them.
Given the reality of frequent human-animal contact, routine characterisation of respiratory infections would seem a sensible precaution to prevent future emergence of Sarbecoviruses.”
All available evidence indicates the requirement of a reservoir host with a wide geographical range, like Chinese horseshoe bats. While the study presents evidence for R. affinis’s importance, there are at least 20 different Rhinolophus species across China. Also, since the sarbecoviruses are generalist in nature, wild or farmed animals such as minks could also facilitate virus transmission from bats to humans.
The risk of future emergence of a new SARS-CoV-2 nCoV strain is too high to restrict sampling strategies,” caution the researchers.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Lytras, Spyros, Joseph Hughes, Wei Xia, Xiaowei Jiang, David L Robertson (2021) Exploring the natural origins of SARS-CoV-2. bioRxiv preprint server.doi: https://doi.org/10.1101/2021.01.22.427830, https://www.biorxiv.org/content/10.1101/2021.01.22.427830v1
- Peer reviewed and published scientific report.
Lytras, Spyros, Joseph Hughes, Darren Martin, Phillip Swanepoel, Arné de Klerk, Rentia Lourens, Sergei L Kosakovsky Pond, Wei Xia, Xiaowei Jiang, and David L Robertson. 2022. “Exploring the Natural Origins of SARS-CoV-2 in the Light of Recombination.” Edited by Adi Stern. Genome Biology and Evolution 14 (2). https://doi.org/10.1093/gbe/evac018. https://academic.oup.com/gbe/article/14/2/evac018/6524630.