Researchers in Sweden have provided a comprehensive characterization of circulating granulocytes in people infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19).
Granulocytes are highly abundant phagocytic white blood cells containing cytoplasmic granules of enzymes that help to digest pathogens.
Now, a team from The Karolinska Institute and Karolinska University Hospital has delineated the contribution of different granulocyte subsets to the pathogenesis of COVID-19.
The findings point to specific immunotypes that could be of value in predicting key clinical features associated with severe disease, say Magda Lourda and colleagues.
A pre-print version of the research paper is available on the medRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Dysregulated inflammatory state as a feature of COVID-19
Since SARS-CoV-2 was first discovered in Wuhan, China, in late 2019, many researchers have proposed that a dysregulated inflammatory state is a significant driver of the clinical complications seen in patients with COVID-19.
Studies have also suggested that dysregulation of the granulocyte family (neutrophils, eosinophils and basophils) plays an important role.
“Accumulating evidence shows that granulocytes are key modulators of the immune response to SARS-CoV-2 infection and their dysregulation could significantly impact COVID-19 severity and patient recovery after virus clearance,” write the researchers.
Clinical characteristics of COVID-19 include increased levels of circulating neutrophils and substantial granulocyte enrichment in the lung.
Longitudinal studies have shown that neutrophil numbers slowly decline throughout the course of infection, but that this rate of decline is slower among severely ill patients.
Interestingly, one study conducted in 2020 found that basophil and eosinophil counts were inversely correlated with neutrophil levels, says Lourda and colleagues.
“However, a comprehensive phenotypic description of circulating granulocytes in SARS-CoV-2-infected patients is lacking,” they write.
What did the researchers do?
The researchers set out to delineate the phenotypic changes in granulocyte populations during the acute and convalescent phases of CVOID-19.
Using high-dimensional flow cytometry, the team performed granulocyte immunophenotyping of peripheral blood and analyzed the diversity of neutrophil, eosinophil and basophil subsets at single-cell resolution.
What did they find?
The team identified specific immune traits in the granulocyte subsets that were associated with the severity of COVID-19.
Severe disease was associated with increased peripheral blood levels of neutrophils and decreased levels of eosinophils and basophils.
“General neutrophilia can be considered a hallmark of severe COVID-19,” writes Lourda and colleagues.
The researchers say the low levels of eosinophils and basophils may result from these cells being recruited to inflamed tissue, particularly the lung.
This hypothesis was supported by the observed altered expression of a number of receptors required for the activation, adhesion and migration of granulocytes, including CD62L, CD11a/b, CD69, CD63, CXCR4.
Furthermore, levels of key soluble factors involved in recruiting both of these cell types (including CCL13, CCL17, CCL22 390 and CCL28) correlated with their levels in circulation.
Paired longitudinal sampling revealed phenotypic restoration of the granulocytic signature during the convalescent phase.
“Our findings are in agreement with previous studies showing the replenishment of the eosinophil and basophil pool and the normalization of neutrophil numbers in circulation,” says the team.
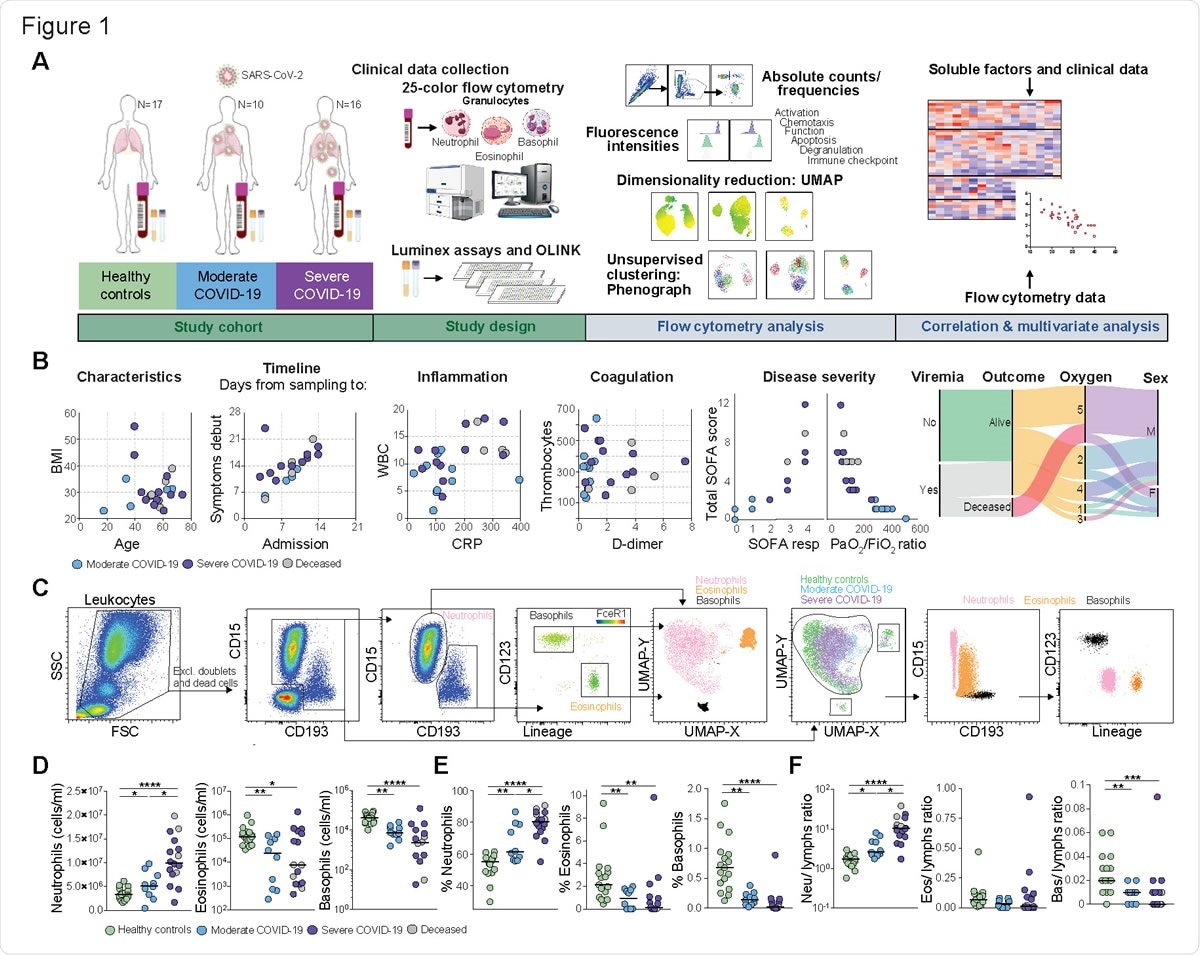
Disease severity-dependent neutrophilia and marked decrease of eosinophils and basophils in COVID-19. (A) Experimental and analytical workflow of the study performed on samples from moderate and severe COVID-19 patients and age-matched healthy controls. (B) Dot plots (left) and alluvial diagram (right) describing demographics and the clinical characteristics of the patients included in the study cohort. Severity groups (moderate=blue; severe=purple; deceased=gray) are indicated. (C) Gating strategy for the identification of granulocyte subsets and their UMAP projection is shown on the left. On the right, analogue results are obtained through unsupervised subset identification based on UMAP projection of total granulocytes. (D-E) Absolute cell counts (D) and frequencies (E) among total leukocytes for neutrophils, eosinophils and basophils in healthy controls (n=17), moderate COVID-19 patients (n=10) and severe COVID-19 patients (n=16). (F) Ratios of granulocyte absolute counts over lymphocyte absolute counts in healthy controls (n=17), moderate COVID-19 patients (n=10) and severe COVID-19 patients (n=16). In (D–F), Kruskall-Wallis test and two-stage Benjamini, Krieger and Yekutieli test. Bars represent median. * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. Neu, neutrophils; eos, eosinophils; bas, basophils; lymphs, lymphocytes.
Immunological parameters helped to predict disease outcomes
The researchers also found that clinical features of disease such as multi-organ and respiratory dysfunction could be predicted using a combination of the immunological parameters and laboratory measurements.
Notably, the immunological parameters were predominant in driving the prediction models, compared with standard laboratory measurements.
More specifically, multi-organ failure and reduced pulmonary function were associated with specific phenotypes, primarily the eosinophil activation markers CD11a, CD66b, and CD69.
“We provide the proof-of-principle that the combination of immunological, and specifically granulocyte-related, measurements with standard clinical data could be used in generating algorithms for patient classification and, thus, tailored therapeutic regimens,” says the team.
What did the authors conclude?
Lourda and colleagues say that overall, the findings extend the current understanding of the distinct contribution of granulocyte subsets to COVID-19 pathogenesis.
“Our findings highlight the significant alterations of granulocyte subpopulations in frequency and function in the blood of patients with COVID-19,” they write.
“Moreover, our data point towards the combined use of granulocyte-related immunological parameters and basic clinical laboratory tests as better prognostic biomarkers of disease severity and disease course,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Lourda M, et al. High-dimensional profiling reveals phenotypic heterogeneity and disease-specific alterations of granulocytes in COVID-19. medRxiv, 2021. doi: https://doi.org/10.1101/2021.01.27.21250591, https://www.medrxiv.org/content/10.1101/2021.01.27.21250591v1
- Peer reviewed and published scientific report.
Lourda, Magda, Majda Dzidic, Laura Hertwig, Helena Bergsten, Laura M. Palma Medina, Indranil Sinha, Egle Kvedaraite, et al. 2021. “High-Dimensional Profiling Reveals Phenotypic Heterogeneity and Disease-Specific Alterations of Granulocytes in COVID-19.” Proceedings of the National Academy of Sciences 118 (40). https://doi.org/10.1073/pnas.2109123118. https://www.pnas.org/doi/full/10.1073/pnas.2109123118.