A recent study conducted at the University of Texas at Austin, USA, has identified a highly conserved neutralizing antibody binding site on the spike S2 domain of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The study findings might help develop novel spike-targeting antibodies for widespread protection. The study is currently available on the bioRxiv* preprint server.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of the coronavirus disease 2019 (COVID-19) pandemic, is a member of the family Coronaviridae. This single-stranded, positive-sense beta coronavirus is closely related to other lethal members of the human coronavirus family, including severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV).
The entry of coronaviruses into host cells is mediated by the spike glycoprotein present on the viral surface. Of two domains (S1 and S2) of the spike protein, the S1 domain contains an N-terminal domain (NTD) and a receptor-binding domain (RBD). Mechanistically, the “up” and “down” conformational states of RBD facilitate receptor binding and immune evasion, respectively. Upon binding of the RBD with the host cell angiotensin-converting enzyme 2 (ACE2), proteolytic activation of the spike protein by host cell proteases takes place, leading to the release of the S1 from the S2 domain and initiation of the virus-host cell membrane fusion.
The majority of the therapeutic monoclonal antibodies are designed to target the RBD-ACE2 interaction. Similarly, plasma samples obtained from COVID-19 recovered patients commonly contain antibodies that target this interaction. However, because the RBD is poorly conserved across different human coronaviruses, antibodies specifically targeting this domain show limited cross-reactivity. In contrast, the spike S2 domain is highly conserved across all human coronaviruses, and thus, can serve as a potential target for neutralizing antibodies.
Study design
The current study focuses on identifying novel antibody binding epitopes on the highly conserved S2 domain of highly pathogenic coronaviruses. The scientists isolated antibodies from mice immunized with S2 protein of MERS-CoV and examined the SARS-CoV-2 spike neutralizing efficacy of these antibodies.
Important observations
The scientists isolated and characterized three highly cross-reactive antibodies (4H2, 4A5, and A3A) that bind to the spike protein of SARS-CoV-2, SARS-CoV, and MERS-CoV with similar efficiency. However, the spike-neutralizing ability was observed only for the A3A antibody. While 4H2 and 4A5 interacted with conserved linear epitopes without any neutralizing effects, antibody A3A showed significant neutralizing effects upon binding to a conformational epitope conserved across all beta coronaviruses. Moreover, A3A showed greater sensitivity for the highly infectious D614G variant of SARS-CoV-2.
The mass spectrometric analysis showed that A3A binding epitope is located at the apex of the S2 domain. Further analysis revealed that the epitope remains entirely shielded by the S1 domain when all RBDs are in the “down” state. In contrast, the epitope is gradually exposed during the progressive transition of the RBDs to the “up” state.
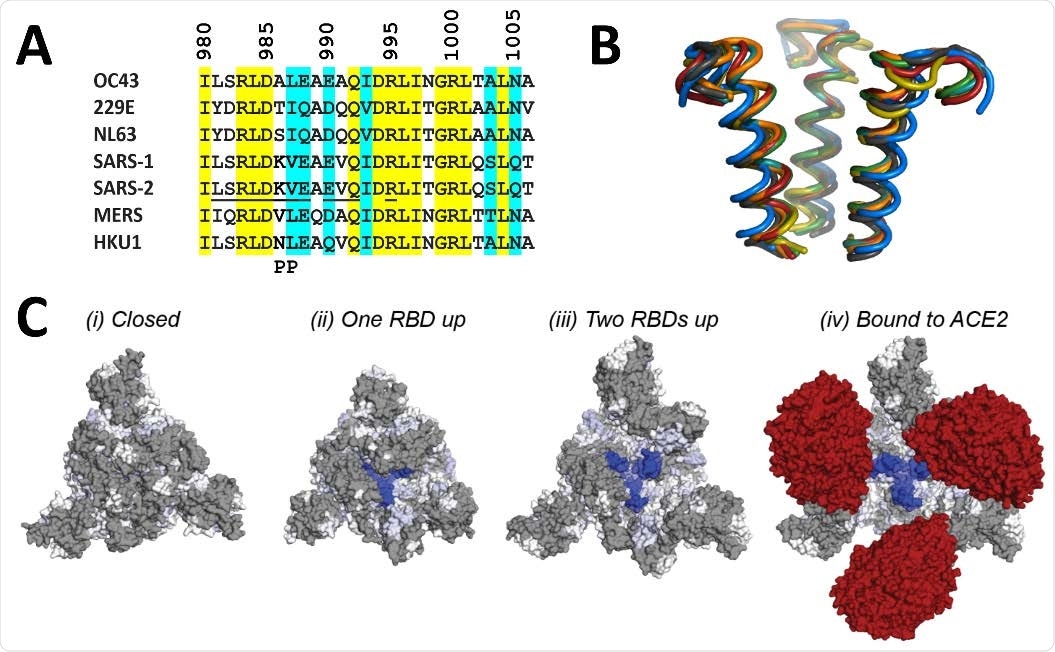
Structural location of 3A3 epitope and implications for antibody binding. The 3A3 epitope identified by HDX mass spectrometry (SARS-2 amino acids 980-1006) is highly conserved across the spike (A) sequences and (B) structures of coronaviruses known to infect humans. In (A), identical residues are highlighted in yellow and similar residues are highlighted in aqua. Solvent exposed residues visible in spike structures with at least one RBD-up are underlined in the SARS-2 sequence. The location of the two proline mutations introduced to 2P variants is shown below the alignment. In (B), the structure of each epitope is displayed as follows: SARS-2 (6VSB) – red, SARS-1 (6CRV, RMSD = 0.8 Å) – orange, MERS (5X5C, RMSD = 3.1 Å) – blue, NL63 (7KIP, RMSD = 2.6 Å) – grey, HKU1 (5I08, RMSD = 0.5 Å) – teal, OC43 (6OHW, RMSD = 0.6 Å) – green, 229E (6U7H, RMSD = 2.0 Å) – yellow. (C) Trimeric SARS-2 spike in various conformations colored according to the difference in deuterium fractional uptake between SARS-2 HexaPro spike alone and with 3A3 IgG (as in Figure 4D). The 3A3 epitope (dark blue) within S2 is completely hidden by S1 in the structure of wild-type SARS-2 spike in the (i) three RBDs down or closed conformation (PDB: 6XR8). In structures of stabilized spike with (ii) one RBD-up (PDB: 6VSB), (iii) two RBDs up (PDB: 7A93), or (iv) three RBDs up and bound to ACE2 (red) (PDB: 7A98), the 3A3 epitope is increasingly accessible. Residues lacking coverage in the HDX experiment are indicated in grey. The side view is shown in fig. S10C.
Regarding virus neutralization, A3A was found to neutralize the D614G variant of SARS-CoV-2 more potently than the wild-type virus. The spike protein of the highly infectious D614G variant is more stable than the spike protein of the wild-type virus, with all RBDs mostly remain in the “up” state. Flow cytometric analysis conducted in the study revealed that A3A readily binds D614G in the absence of the receptor and potently neutralizes pseudoviruses expressing spike protein with D614G mutation. Despite not being able to assess the epitope on the wild-type spike readily, A3A could still neutralize the wild-type spike. This indicates that the antibody can bind and neutralize the wild-type spike after its interaction with ACE2.
The epitope of A3A was found to be highly conserved (70% sequence similarity) across all human coronaviruses. The antibody showed similar binding sensitivity for the spike protein of SARS-CoV-2, SARS-CoV, and MERS-CoV, which indicates that the binding ability of A3A depends only on the RBD conformational states and not on the RBD sequence.
Study significance
The study identifies a novel neutralizing epitope on the S2 domain of SARS-CoV-2 spike protein, which has high sequence identity and similarity among all human coronaviruses. The A3A antibody binds and locks the spike protein in pre-fusion conformation, indicating that the antibody can be used to evaluate spike quality. Notably, the novel epitope becomes more available to A3A antibodies in viral variants that stabilize the RBD in an “up” conformational state, which is a basic feature of highly transmissible viruses. Taken together, the A3A epitope on the S2 domain can serve as a potential target for developing highly potent therapeutic antibodies and vaccines.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
bioRxiv preprint server. 2021. Huang Y. Identification of a conserved neutralizing epitope present on spike proteins from highly pathogenic coronaviruses. https://www.biorxiv.org/content/10.1101/2021.01.31.428824v1
- Peer reviewed and published scientific report.
Silva, Rui P, Yimin Huang, Annalee W Nguyen, Ching-Lin Hsieh, Oladimeji S Olaluwoye, Tamer S Kaoud, Rebecca E Wilen, et al. 2023. “Identification of a Conserved S2 Epitope Present on Spike Proteins from All Highly Pathogenic Coronaviruses.” Edited by Leslie Goo and Sara L Sawyer. ELife 12 (March): e83710. https://doi.org/10.7554/eLife.83710. https://elifesciences.org/articles/83710.