According to the World Health Organisation (WHO), the coronavirus disease 2019 (COVID-19) has to date infected more than 102 million people and claimed over 2.25 million lives globally. Several vaccines are now available. However, some experts have suggested that longer-term endemic infection levels may persist, with intermittent waves of resurgence when circulating strains acquire new pathogenic mutations evading vaccine-stimulated immunity.
An effective COVID-19 response will likely require widespread vaccine distribution and a complete picture of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pathogenic mechanisms.
New research suggests the modulation of lysosomal-immune pathways may present a novel drug-targeting strategy to attenuate SARS-CoV-2 infections.
The recent paper posted on the bioRxiv* server makes use of the published single-cell RNA-seq data obtained from clinically staged COVID-19 patients. Analyzing this data, Rahul Pande and team, from Translational Sciences, Sanofi, USA, profiled COVID-19 responses and identified pathway activities associated with moderate versus severe clinical phenotypes. They found that lysosomal-immune axis signaling is significantly associated with COVID-19 severity.
In this study, they identified a protein interactome for cytokine signaling associated with severe COVID-19 cases. From their results, they observe distinctive transcriptomic profiles for moderate versus severe COVID-19 cases - with the lysosomal-immune system pathways in COVID19 pathogenesis offering novel drug targets.
This study was undertaken using single-cell RNAseq data obtained from cells in bronchoalveolar lavage fluid (BALF) samples from Healthy Controls (HC; n = 3 patients; 20991 cells) and from COVID-19 patients clinically staged as either Moderate (M; n = 3 patients; 8084 cells) or Severe (S; n = 6 patients; 46001 cells).
Single-cell RNA sequencing (single-cell RNAseq) is a powerful technology that allows profiling the whole transcriptome of a large number of cells. It generates large volumes of data. Processing and analysis of the extensive scRNA-seq data require specialized statistical and computational methods.
For analyses, the researchers in this study used the online freely downloadable R packages: Seurat (https://satijalab.org/seurat/install.html) and WGCNA (https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/). The other tools and the detailed protocols employed in the study are elaborated in the paper.
The researchers performed transcriptomic profiling to identify broad cell types involved in infection response. This annotation identified numerous macrophages along with other immune cell types in all samples. However, they observed the disease severity-associated macrophage populations in severe COVID-19 samples.
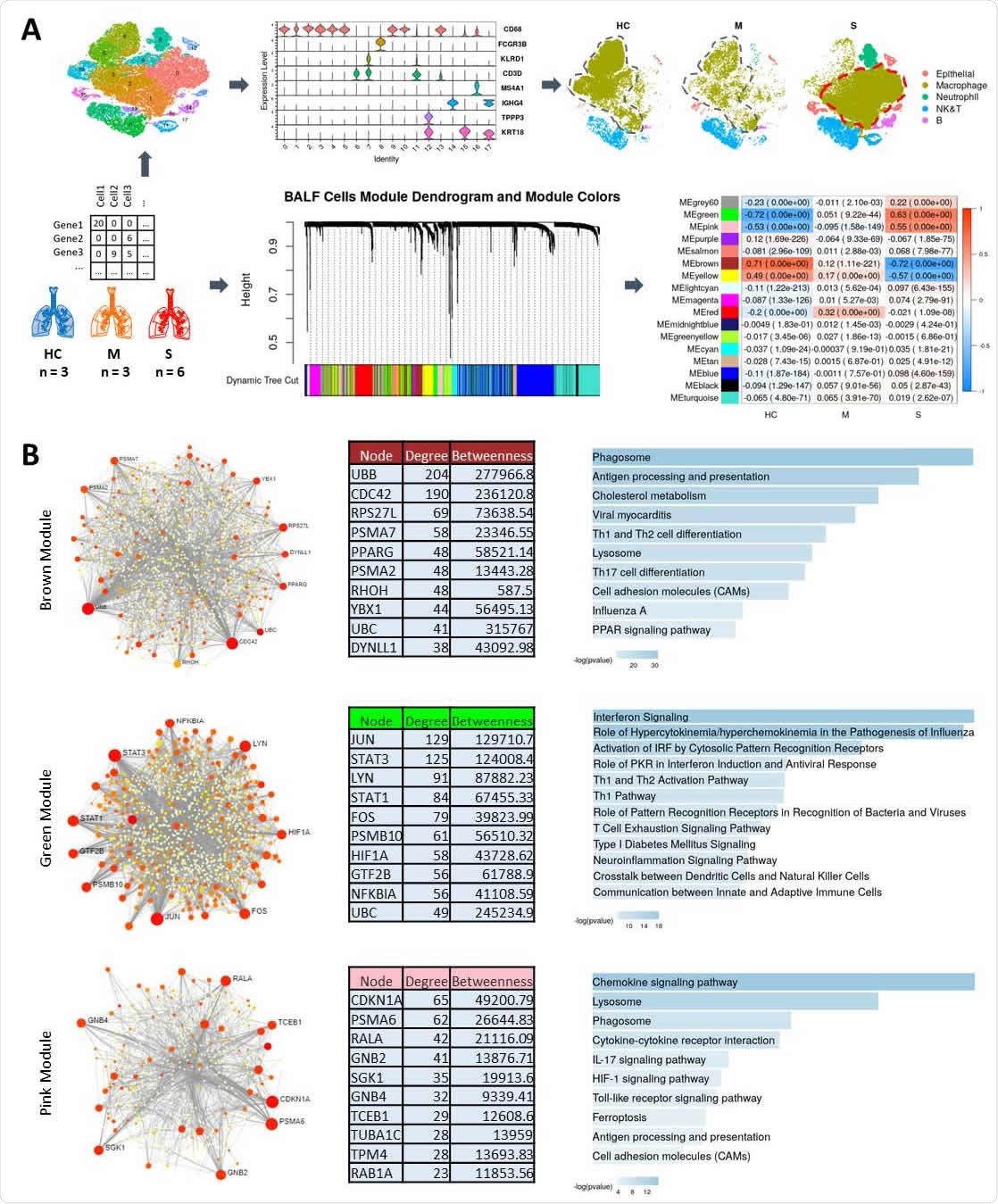
Transcriptomic Profiling and Weighted Gene Co-Expression Analysis (WGCNA}. Schematic of the workflow showing single cell data from Healthy Control (HC), Moderate (M), and Severe (COVID-19) cases used for broad cell types clustering and WGCNA to identify modules significantly correlated with disease severity. Also, note the change in clustering and separation of macrophage portion in Severe cases. (A). PPI networks and pathway functional enrichments for genes in highly correlated modules are shown for Brown, Green, and Pink modules (B}.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
They identified gene co-expression network disease- and severity-associated modules - they present 17 discrete co-expression modules. Out of these, two modules (represented as green and pink) were found to be positively correlated with the severe disease. The significant and robust correlation indicates the upregulation of pathways linked with these modules among critically ill patients.
The genes in the protein-protein interaction (PPI) network associated with one of the modules (green) are enriched for pathways involved in the "cytokine storm." The PPI network for the Green module shows STAT3, STAT1, JUN and FOS as hubs.
Likewise, the genes in the other module (pink) are highly enriched for chemokine signaling, IL17 signaling, and lysosomal compartment.
"These module expression patterns corroborate earlier findings showing that alveolar macrophage may be the main driver of 'cytokine storm' in the severe patients [25, 26] and the observation suggests that macrophage-specific drugs targets could be crucial for an effective therapy for COVID19."
The researchers present a heatmap of the observations where the gene expression splits between the healthy controls and the severe disease cases.
Interestingly, they found that lysosomal-immune axis signaling is significant with COVID-19 severity - evidence from previous and current studies corroborate this observation. The lysosomal-immune system pathways in COVID19 pathogenesis may offer novel drug targets.
The results here suggest distinctive transcriptomic profiles for moderate versus severe COVID-19. They also identified a protein interactome for cytokine signaling associated with severe COVID-19 cases. The study also identifies a new gene co-expression and PPI networks that may mediate disease severity.
This study provides new support and clinical validation for these observations on the host-virus interaction's underlying mechanism. The researchers write these findings have implications for the new drug development and existing drug repurposing against COVID19.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.