With new variants emerging, often showing increased transmissibility and virulence, governments and health organizations do not appear to be within striking distance of effectively containing the ongoing coronavirus disease 2019 (COVID-19) pandemic. Caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), this condition presents with a wide spectrum of severity, ranging from asymptomatic infection through mild or moderate COVID-19 to severe or critical disease.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Viral sgRNA production
Following SARS-CoV-2 infection of the host cell, the single-stranded virus uses the cell machinery to set up its replication-transcription complex. Within the cell, both viral genomic replication and transcription of subgenomic transcription occur.
The first results in the production of 30 kb genomic RNA (gRNA), while the second process causes a distinct set of spliced subgenomic RNA transcripts (sgRNA) to be produced by discontinuous transcription. These sgRNAs act as viral mRNAs, in order to initiate the translation of many structural and accessory proteins that are essential for the production of new viral particles, their packaging and release.
The presence of sgRNA occurs within infected cells, but it is obvious that only gRNA is packaged into the new virion. Thus, sgRNAs could be a proxy for viral replication and for viral fitness within the host.
Disease characteristics and sgRNA
Recent studies show that structural variations in sgRNAs affect the severity of the disease, transmissibility and immune response. The current study showed clear links between the type and expression of sgRNA and the clinical severity of COVID-19.
Different types of sgRNA retained the same proportion and expression ranking in each sample. In all samples, the sgRNA encoding the nucleocapsid or N protein was most abundant, and that transcribing ORF7b the least.
Deletions distinguish symptomatic infection
The researchers also found distinct sgRNA deletion sets in symptomatic vs. asymptomatic infections. Such deletions were widespread. The reason suggested is the increased viral replication in symptomatic infection, leading to a higher generation of structural variants.
Of over 8,500 unique deletions found in two or more independent reads, 6% (~500) of these deletions occurred in a tenth of samples at least. Among these high-frequency deletions, 375 and 38 were specific to symptomatic and asymptomatic hosts, respectively, while 88 were found in both types. However, those occurring only in symptomatic hosts were both more abundant and larger at ~200 nucleotides, vs. 46 in asymptomatic hosts.
This could indicate, they say, “a potential selection force for different types of viral variants adapted in distinct cohorts of host responses.”
ORF3a deletion
When the deletions were examined by frequency of symptomatic vs. asymptomatic samples, almost 300 deletions were preferentially found in symptomatic infections, but only 10 in asymptomatic. Of these, 263 vs. 9 deletions were specific to symptomatic and asymptomatic samples, respectively.
Of the ten deletions found mostly in asymptomatic infection, three were in the sgRNA protein-coding regions. Two of these three were in the ORF3a sgRNA.
The ORF3a protein is concerned with inducing infected cells to enter apoptosis (or programmed cell death), thus regulating the inflammatory response following infection.
These deletions would cause truncation of the ORF3a protein, with weaker pro-apoptotic properties, and mild inflammation, thus resulting in very mild or asymptomatic infection.
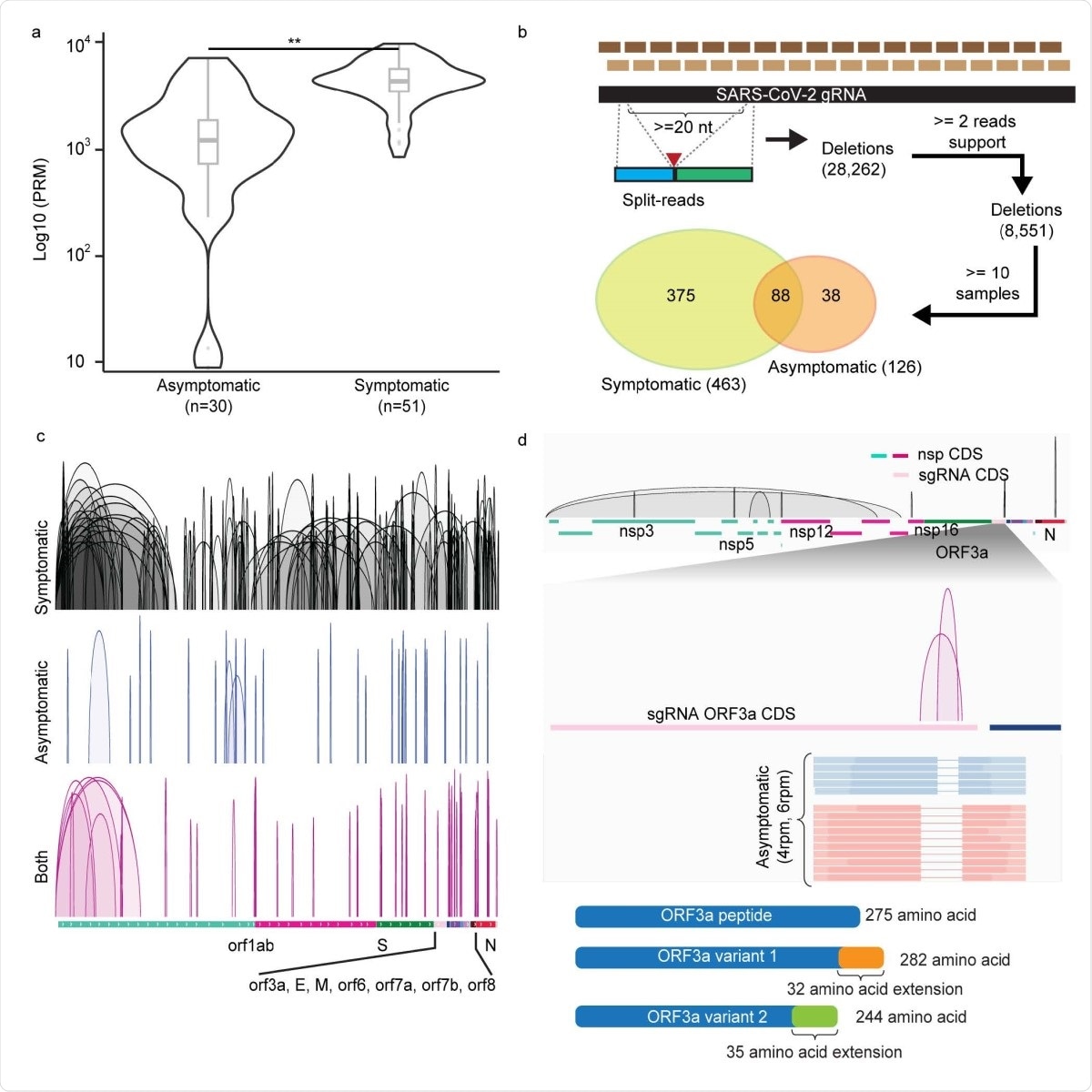
Deletions of SARS-CoV-2 RNAs in symptomatic and asymptomatic COVID-19 positive patients. a. Distributions of normalized split-aligned reads counts in asymptomatic and symptomatic patients. Two-sided Wilcoxon Rank-Sum Test, p = 2.3 × 10-8. Center line, median; boxes, first and third quartiles; whiskers, 1.5 × the interquartile range. b. Deletions inferred by amplicon-seq data from asymptomatic and symptomatic patients’ specimens. c. Visualization of the deletions detected in symptomatic (n=287), asymptomatic (n=34) and both (n=79) samples in IGV genome browser in reference annotated subgenomic RNA (sgRNA) transcribed regions. d. Top: Deletions (n=10) preferentially found in viral RNAs from the asymptomatic samples. Middle: zoom-in view in sgRNA_ORF3a coding sequence (CDS) region shows the two deletions uniquely found in asymptomatic cases, their normalized counts and representative read supports. Lower: their predicted translated peptide in reference to the wildtype ORF3a peptide.
Functional changes linked to deletions
Similar associations with asymptomatic infection were found in the case of deletions in the protein-coding region of sgRNA for other major structural and non-structural proteins encoding the N protein and essential viral enzymes.
The researchers point out that the presence of distinct deletions in the viral sgRNAs in infected symptomatic and asymptomatic individuals, consistently observed across a range of different hosts, strongly supports their contribution to altered function within the different structural variants. Such changes in function could lead to altered virulence and pathogenicity between variants.
Deletions arise during replication
The wide range of deletions is notable. In order to understand their origin, whether during replication or transcription, and how they affected the protein products, the researchers performed full-length sequencing in order to explore them against the genetic background of the sgRNA structure as a whole.
They found that most derived from poor RNA polymerase fidelity leading to inaccurate viral gRNA replication. These were then incorporated via transcription into the sgRNAs. Such structural variations are likely to be very frequent and may be present in a large percentage of the viral population in a host, during active replicative infection.
The presence of a number of alternative sgRNAs and the protein versions they encode could indicate the presence of quasispecies within a host. This could accelerate adaptation to the host, a phenomenon often observed in other RNA viruses.
Protein variants may affect therapeutic efficacy
Most of the encoded proteins among the sgRNA variants from these clinical samples were full-length proteins. The least affected were ORF6 and envelope protein, predicted to encode wildtype full-length protein in over 90%. However, the most common variants of the spike and of ORF3a proteins were those with the now globally dominant D614G mutation, and the Q57H mutation, which has increased in proportion since February 2020.
As a result of such sgRNA variants, five types of encoded proteins were found; namely, wildtype annotated proteins, annotated proteins with substituted amino acids, annotated truncated proteins, annotated proteins with C-terminal extensions, and new peptides.
Among the truncated protein products, 56% and 41% of the spike and N proteins were included. When annotated, the researchers found that, surprisingly, over 40% of the truncated spike and N proteins lacked the receptor-binding domain (RBD) and RNA-binding, respectively. The loss of these protein domains would prevent viral entry into the host cell, and virus transcription and assembly, respectively.
Since the spike and N proteins are major targets for developing vaccines, therapeutic antibodies and antiviral drugs, their loss or functional truncation could have serious implications on the efficacy of such strategies.
What are the implications?
The researchers found that viral genomes from asymptomatic and symptomatic infections contain distinct arrays of deletions. Each deletion array is preferentially and reliably linked to one of these presentations only. This suggests that sgRNA profiles are significantly linked to the clinical features of the infection.
A closer examination of the predicted viral proteomes, using full-length viral transcripts as the base, showed that SARS-CoV-2 is undergoing significant and rapid change.
With asymptomatic infection, there is a relative reduction in sgRNA production, indicating repression of viral transcription independent of the viral load. As such, sgRNA characterization of PCR-positive samples could help distinguish those with active replication from others.
Notably, the sgRNA/gRNA ratio bears a strong correlation with clinical symptoms. As such, “RT-qPCR based assays to quantitatively evaluate the relative abundance of sgRNAs may be a predictive measure of the clinical severity of COVID-19 symptoms.” This could help allocate healthcare resources where urgently required in scenarios where they are under intense pressure due to spiking cases and mortality rates.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.