A recent study conducted by US scientists has demonstrated that the distribution of local topological free energy along the spike protein domains of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is associated with their activity in protein rearrangement.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The study also reveals that spike protein conformation with high local topological free energy are associated with mutations that significantly affect protein rearrangement. The study is currently available on the bioRxiv* preprint server.
Background
Coronavirus disease 2019 (COVID-19) cause by SARS-CoV-2 has already claimed more than 2.3 million lives globally since its emergence in December 2019. To effectively control SARS-CoV-2 transmission, it is important to understand the dynamics of viral infection cycle. Based on scientific literature, the interaction between SARS-CoV-2 spike protein and host angiotensin-converting enzyme 2 (ACE2) and the fusion of viral envelope with host cell membrane are the key events to establish an infection. The spike protein's activation is initiated by proteolytic cleavage at the S1 subunit, followed by a second cleavage at the S2 subunit, leading to cell-cell fusion and viral entry. For the entire process of viral entry, multiple rearrangements in the spike protein structure is a prerequisite.
In the current study, the scientists aim to investigate how local rearrangements in spike protein can induce global conformational changes. To characterize various spike protein conformations at the length-scale of 4 consecutive residues along the protein backbone, they have used tools (the Writhe and the Torsion) from knot theory, which is a mathematical study of three-dimensional arrangements of closed curves (knots). By using the Writhe and the Torsion, they have introduced a new local topological free energy along the viral protein backbone to investigate the involvement of different local topological free energy conformations in protein folding.
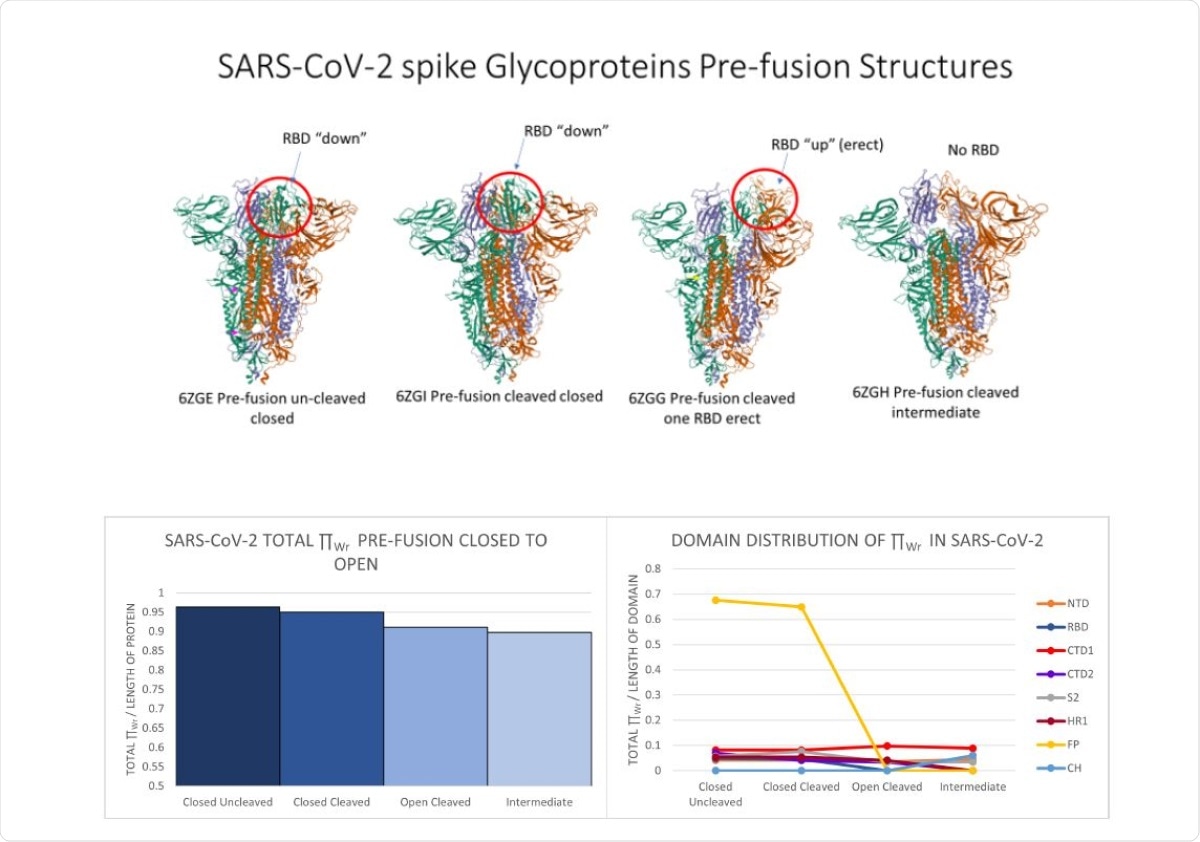
Top: From left to right, snapshots of SARS-CoV-2 pre-fusion protein at four stages: uncleaved closed (6ZGE), cleaved closed (6ZGI), cleaved open (6ZGG) and intermediate (6ZGH). The RBD is circled in red. Bottom Left: The normalized total local �Wr for SARS-CoV-2 protein at the 4 pre-fusion stages. Bottom Right: The normalized total local �Wr-values for SARS-CoV-2 protein domains at the 4 pre-fusion stages. Crystal structure images were pulled from the Protein Data Bank.
Important observations
According to the findings, the total local topological free energy reduced from pre-fusion to post-fusion for all representative viral proteins tested. This indicates that during fusion, viral proteins are directed toward a minimal total local topological free energy state. The same feature was also observed for SARS-CoV-2 spike protein. Moreover, a continuous reduction in the total local topological free energy of spike protein was observed in different conformational stages (uncleaved closed, cleaved closed, cleaved open and intermediate state) that ultimately lead to protein rearrangement. This indicates transition to a more energetically stable state. This was further supported by the observation that SARS-CoV-2 harboring G614 had significantly higher total local topological free energy than SARS-CoV-2 harboring D614. There is evidence suggesting that G614-containing viral variant is much more unstable than those containing D614.
Moreover, an alteration in distribution of the local topological free energy in different spike protein domains was observed for closed and open and pre-fusion and post-fusion states. These observations suggest that high total local topological free energy in spike domains can specify their activity in protein arrangement.
The results obtained from density functional theory calculations showed that local spike protein conformations with medium or low local topological free energy reduced to conformations with even lower local topological free energy. In contrast, local conformations with high local topological free energy reduced to conformations with both lower and higher local topological free energy. This further indicates that protein conformations with high local topological free energy are more unstable.
By comparing mutations known to increase or hinder spike protein rearrangement and stability, it was observed that most of the mutations so far known to increase the transmissibility and virulence of SARS-CoV-2 had occurred at residues present in conformations with high local topological free energy.
Taken together, the study reveals that SARS-CoV-2 spike protein conformations with high local topological free energy are more unstable than those with low local topological free energy. Moreover, the study indicates that spike conformations with high local topological free energy are associated with mutations that are known to affect protein rearrangement.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources