Even as vaccination is being rolled out against the coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), scientists grapple with the threat of new and more transmissible variants, which may be resistant to the antibodies elicited by either natural infection or vaccination. This would severely endanger the achievement of population immunity, the goal of vaccination, and the hope of a return to everyday pre-pandemic life.
Study: Naive human B cells can neutralize SARS-CoV-2 through recognition of its receptor-binding domain. Image Credit: Design_Cells / Shutterstock
Adaptive immunity

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
When an individual is exposed to a viral antigen, both an immediate and a delayed anamnestic immune response occur. The human immune system contains potential B cell receptor (BCR) rearrangements that can recognize even newly encountered antigens, triggering these naïve B cells' activation.
This, in turn, leads to affinity maturation via a process of somatic hypermutation and differentiation. Affinity maturation refers to the process by which activated B cells produce antibodies that have higher affinities for the antigen over the course of the immune response. The outcome is often the production of neutralizing antibodies to the pathogen.
Memory B cells induced by exposure to SARS-CoV-2 may persist in the host, and affinity maturation has been seen to continue for more than six months after primary infection, indicating that this exposure could provide long-term protection against reinfection. Comparable levels of neutralizing antibody titers have been measured in vaccine recipients as in recovered COVID-19 patients.
SARS-CoV-2 RBD
SARS-CoV-2 binds to the host cell receptor, the angiotensin-converting enzyme 2 (ACE2), via the receptor-binding domain (RBD) on the viral spike glycoprotein. Many neutralizing antibodies bind the RBD and thus create steric hindrance to the spike-ACE2 binding. An examination of monoclonal antibodies against the RBD shows that they are generated by various heavy- and light-chain variable gene segments.
In other words, the B cell repertoire contains a vast number of potential combinations capable of producing specific anti-RBD antibodies. Thus, the RBD is the major antigenic determinant of the neutralizing response simply because there is a significant frequency of naïve BCRs that already have some affinity for it, indicating they can be easily activated by exposure to the virus.
Thus, this protective response is already present but requires to be amplified by exposure to the antigen via natural infection or vaccination.
The current preprint deals with the antigen specificity of such naïve B cells against the SARS-CoV-2 RBD in terms of their frequency, affinity, and potential to generate a protective response following exposure.
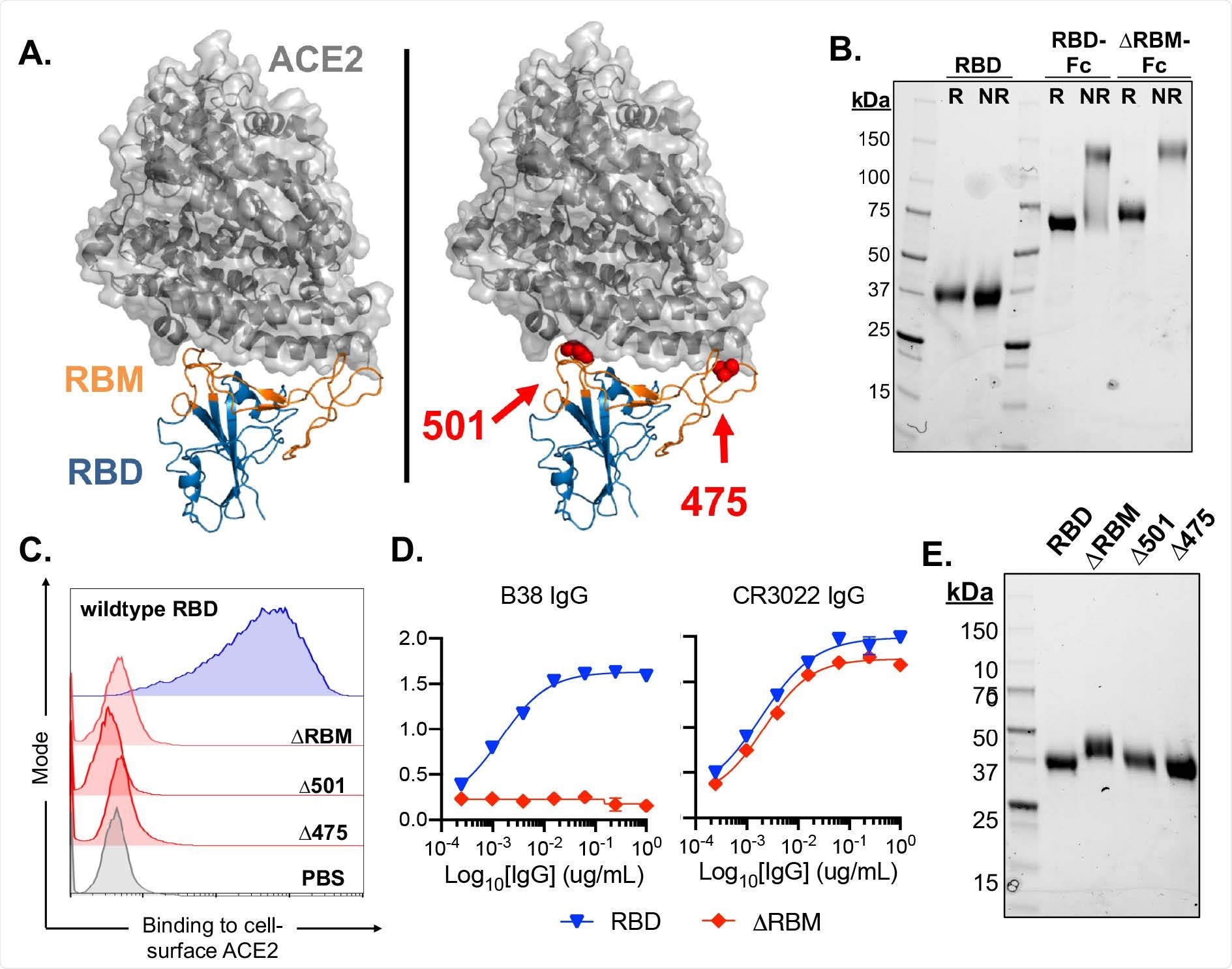
Design and characterization of SARS-CoV-2 antigens. (A) SARS-CoV-2 RBD in complex with viral receptor, ACE2 shown in blue and grey, respectively (PDB 6M0J). Wild-type RBD with, the receptor binding motif (RBM), shown in orange (left panel). Structural model of the ΔRBM probe designed to abrogate binding to ACE2 (right panel). Putative N-linked glycosylation sites engineered onto the RBM are shown in red spheres at amino acid positions 501 and 475. (B) SDS-PAGE gel under reducing (R) and non-reducing (NR) conditions for monomeric RBD, RBD-Fc and ΔRBM-Fc. (C) Wildtype RBD, ΔRBM and single glycan variant binding to ACE2-expressing 293T cells by flow cytometry. Wild-type RBD binding shown in blue, glycan variant binding shown in red. Streptavidin-PE was used to detect the relative intensity of antigen binding to cell-surface ACE2. A PBS control (gray) indicates secondary-only staining. (D) Control antibody ELISA binding to RBD and ΔRBM antigens. RBM-specific antibody, B38 (left). Non- RBM-specific control antibody, CR3022 (right). (E) ΔRBM and Δ501 and Δ475 variants analyzed by SDS-PAGE gel under reducing conditions; wildtype RBD is shown for comparison.
Naïve B cells specifically recognize RBD
The researchers found that blood samples from eight seronegative donors contained naïve B cells that specifically bound the viral spike and the RBD, particularly the receptor-binding motif (RBM). They observed that these cells made up about 0.0021% and 0.0023% of total and naïve B cells, respectively. However, they made up 3.2% of naïve spike-reactive cells. This indicates that most of the spike epitopes recognized by naïve B cells lie outside the RBD.
Gene diversity
Analysis of light- and heavy-chain antibody sequences from five of the eight donors showed diverse gene use, with no discernible preferences for gene pairing. This led to the generation of multiple unique antibody clones, as has been observed in RBD-specific memory B cells isolated from convalescent COVID-19 subjects and vaccine recipients.
Thus, antigen exposure leads to the activation of a variety of different antibody precursors. IGHV3-9 genes were increased in frequency to about 20% in these five donors, mirroring the pattern observed in SARS-CoV-2 exposed individuals.
The researchers found enriched IGHV3-53 and 3-30 gene segments in the remaining three donors, typical of anti-RBD antibodies from COVID-19 convalescents.
Overall, there was a wide range of third complementarity-determining regions (CDR3) lengths for both light and heavy chains. Also, the variable heavy and light chains were mostly identical with the germline sequences.
Binding of original and variant SARS-CoV-2
They also measured the binding affinities of the naïve antibodies to SARS-CoV-2 RBD in about 80% of antibodies obtained across five donors. These antibodies could also bind the South African SARS CoV-2 variant 501Y.V2 in about half of the cases (22 antibodies), with one of them binding specifically to this variant but not to the original strain.
While 86% of antibodies were specific to the SARS-CoV-2 RBD, five antibodies cross-reacted to both SARS-CoV and WIV-1, closely related coronaviruses.
Neutralizing potential
Neutralization assays showed that five of the 36 anti-RBD antibodies tested were capable of detectable neutralization. These are less potent, but nonetheless, the presence of neutralizing capacity at germline is noteworthy. Therefore, they can undergo affinity maturation rapidly following vaccination or natural exposure to the virus to form potent RBD-targeting neutralizing antibodies.
These naïve antibodies bind to sites across the RBM, as shown by earlier studies using various methods, including epitope mapping and deep mutational scanning. This could be of great benefit in protecting the individual against different variants of the virus.
What are the implications?
"These data demonstrate that coronavirus-specific naive antibodies are present in distinct donors, are of unrestricted gene usage and when expressed as IgGs, have detectable affinity to RBDs from the currently administered vaccines, a circulating variant of concern, and from at least two related viruses."
These findings clearly distinguish the protective response against SARS-CoV-2 from that against HIV, for instance. Here, not only do germline antibody precursors against the receptor-binding site show low binding affinity, but they have restricted gene usage and cannot neutralize the virus at the germline. In contrast, the germline-specific responses against SARS-CoV-2 offer the potential for the development of broad-spectrum antigens.
"Future investigations aimed at understanding the naive repertoire with respect to coronaviruses of pandemic potential may reveal further commonalties in antigen-specific precursors, enabling the development of pan-coronavirus vaccines aimed at priming specific germline responses of protective potential."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Feldman, J. et al. (2021). Naive human B cells can neutralize SARS-CoV-2 through recognition of its receptor binding domain. bioRxiv preprint. doi: https://doi.org/10.1101/2021.02.02.429458. https://www.biorxiv.org/content/10.1101/2021.02.02.429458v1
- Peer reviewed and published scientific report.
Feldman, Jared, Julia Bals, Clara G. Altomare, Kerri St. Denis, Evan C. Lam, Blake M. Hauser, Larance Ronsard, et al. 2021. “Naive Human B Cells Engage the Receptor Binding Domain of SARS-CoV-2, Variants of Concern, and Related Sarbecoviruses.” Science Immunology 6 (66). https://doi.org/10.1126/sciimmunol.abl5842. https://www.science.org/doi/10.1126/sciimmunol.abl5842.