As efforts continue to unravel the pathogenesis of coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a new study by researchers at the University of California Santa Cruz, USA, discusses the use of simultaneous sequencing of both human and viral genes. This allows the interdependent expression of these transcriptomes to be examined.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Dual RNA-seq explores host-viral transcriptomes
The approach, called dual RNA-seq, seeks to understand how different genes in the host and virus are expressed in an interrelated manner in COVID-19. It involves the examination of both human and viral RNA transcripts simultaneously, in the same sample.
Previously, transcriptional pathways have been studied only in the human host, profiling the differentially expressed genes (DEGs) in cells infected with SARS-CoV-2. Dual RNA-seq examines co-expressed genes, and can identify rare transcript classes, as well as more nuanced associations between genes.
Both infected cells in culture, and patient samples, were analyzed quantitatively for transcripts. With modern sequencing methods, the depth of each read is adequate to allow accurate quantification of the whole host and virus transcriptome without the need for auxiliary library enrichment steps. The method yielded a sensitive and specific network of associations, that showed important roles played by human genes and pathways in SARS-CoV-2 infection.
Comparison with earlier data on SARS-CoV-2 infection
The dual RNA-seq analysis pipeline (dRAP) study first examined data from an earlier study of two infected human cell lines, ACE2 receptor-enhanced alveolar basal epithelial cell line (A549) and bronchial epithelial cell line (NHBE). The amount of SARS-CoV-2 in the cell lines was controlled.
The re-analysis by dRAP showed agreement with the specific transcripts detected. However, since it quantified the amount of viral transcript in the host cell, the researchers found a dramatic shift in the magnitude of the changes within the A549 cell line, compared to the NHBE. This might be because the latter was supposed to be mock-infected but had actually been contaminated with the virus.
This difference in the findings validates the reliability of this method in exploring the changes in the regulation of co-expressed host and viral genes in an infected cell.
In both cell lines, the viral genes that are basic to infection are most highly expressed, such as the open reading frame (ORF) 10, spike, nucleocapsid and membrane genes. Eight of eleven viral transcripts were found in both cell lines, indicating this method can reliably identify viral genes in multiple cell types.
Detection of transcripts in human cells with known infections
The researchers then used the same method to study viral transcript amounts in human cells infected with known infections. The aim was to distinguish human cell transcriptional changes that directly reflect viral transcriptional alterations from those which are indirect effects of the expression of other genes.
They found, unexpectedly, that lung and peripheral blood samples often failed to show any viral gene expression at all, despite the lung alveolar cells being the primary target of the virus. On the other hand, bronchoalveolar lavage fluid (BALF) samples consistently showed overexpression of viral RNA, often in excess of that observed in infected cells in culture.
The most highly expressed viral transcript in BALF was ORF1ab, greatly exceeding that found in infected cell lines. The next in line was the S (spike) protein, followed by the N and M genes. Other genes that were found in excess in BALF samples relative to cell lines were the E, ORF6, and ORF7b genes.
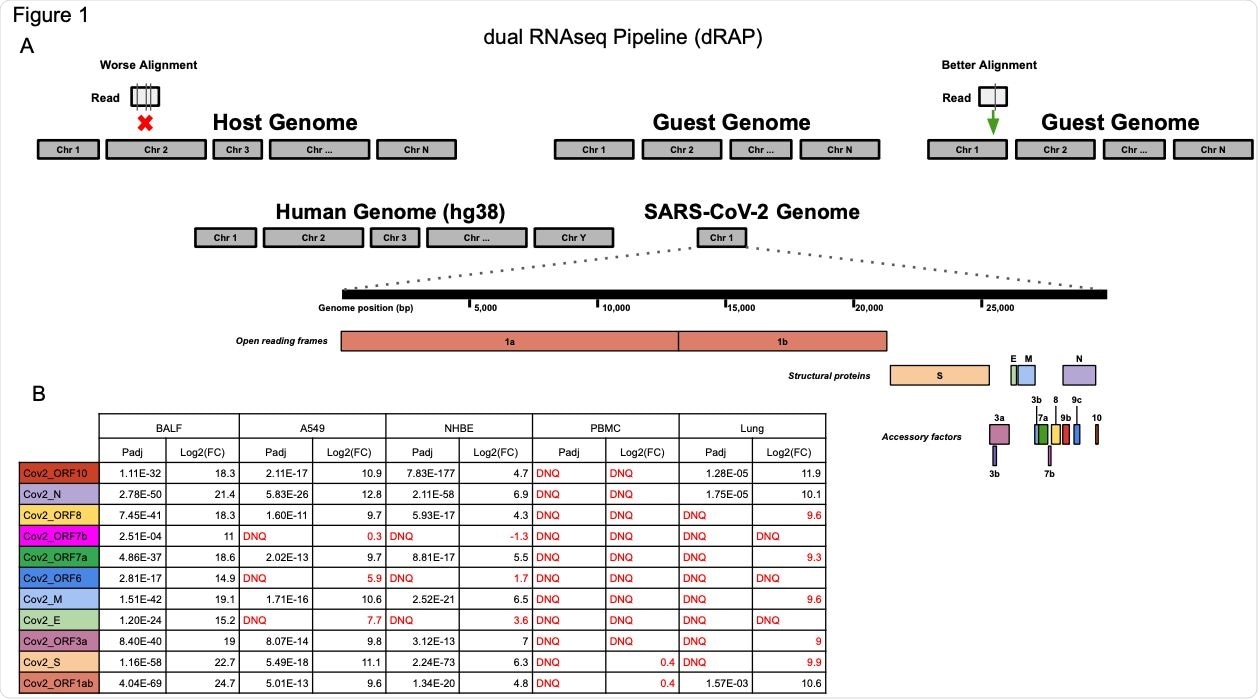
(A) The dRAP pipeline follows a dual RNAseq methodology by creating a single reference index for RNAseq read alignment with STAR by appending “guest” genomes to the host genome. This allows for simultaneous, unbiased alignment of reads with multiple transcriptomes where the best overall alignment is selected. For example, aligning a single read would result in 3 mismatches (vertical lines in box under “Worse Alignment”) if aligned to the host genome (red “x” marks host alignment location) compared to 1 mismatch (vertical line in box under “Better Alignment”) aligning to the guest genome (green arrow). Both the human host genome (hg38) and the SARS-CoV-2 guest genome (NC_045512v2) are included, which has a set of genes that are designated as open reading frames, structural proteins, or accessory factors. (B) SARS-CoV-2 differential gene expression results for infected patient tissue and cell line samples compared with non-infected samples. Patient tissue types show dramatically different expression profiles with PBMC and Lung biopsy tissue rarely ever passing detection limits while BALF tissues show robust expression in infected patients. Cell lines also display strong SARS-CoV-2 expression although the magnitude of fold change was far less than that observed in BALF samples. “DNQ” stands for “did not qualify”, which indicates genes that failed Cook’s distance filtering in DESeq2 analysis.
BALF and cell lines show concordance
Overall, cell line and BALF transcripts showed concordance, except for the overexpression of some viral transcripts in the latter.
Infected A549 cell lines showed similar but lower expression profiles to BALF samples. BALF transcripts were discordant with those in peripheral blood mononuclear cells (PBMCs), which failed to reliably reflect the virus's presence.
The researchers used BALF as well as the two cell lines for further host-virus analysis via DEGs in the former. They postulated that these could reflect direct effects of infection better than lung cells or PBMCs.
There was significant concordance in the gene expression in the two cell lines, 23 and 51 genes in the NHBE and A549, respectively, showing changes in the same direction of expression (mostly upregulation), but with fold differences in the amount. Genes involved in the antiviral response played a large part in the DEG profile.
When BALF DEGs were compared with the two cell lines, both concordant and mismatched directional expression changes were observed.
However, when BALF DEGs were compared with lung and PBMC samples, respectively, most changes tended to show discordance with the former, but extreme discordance with the latter. This confirms the earlier discordance in viral detection between BALF and the lung cells/PBMC.
Human transcriptional network
The viral and host RNA study via coexpression analysis was found to complement DEG analysis by exposing less obvious shifts in the transcription of human genes in sync with viral genes, possibly due to regulatory effects.
Using three different approaches, they found that the gene expression profiles were highly comparable for chemokines, SPRRs, S100s, viral response, and interferon response genes. One result was the generation of a highly modular structure, with specific groups of genes being localized in a group of pathways. This is helpful to distinguish subsets of genes that act together in a specific pathway.
One such example is the pathway involved in a lipopolysaccharide response that is related to high cytokine and chemokine activity, which was found to be linked to another gene cluster regulating epithelial cell differentiation and keratinization, as well as one modulating antiviral host responses.
Ranking analysis showed that many viral genes were highly networked among the DEGs in the cell lines, but not in BALF. This could be due to the over 20-fold increase in the number of DEGs in the former but not in the latter. The increased noise would hide the regulatory interaction network between the genes, especially when interindividual variability is accounted for.
This suggests that many of the DE genes in BALF samples may be more indicative of patient variability than of a response to SARS-CoV-2 infection.”
Consensus network modules
With all three approaches as well as DEG analysis, they showed several genes in common, including five viral genes and 14 host immune genes. Consensus network construction demonstrated several modules, comprising interlinked genes as well as pathways.
These modules consisted of pathways involving interactions between viral proteins and host cytokine and chemokine signaling pathways, and lipopolysaccharide responses, indicating host immune response and disease mechanisms; innate immune pathways, including interferon responses; pathways involving programmed cell death, epithelial differentiation and cornification, and antiviral responses; and viral genome replication and type I interferon-induced response.
These include conventional components such as inflammation and chemokine signaling, as well as unexpected pathways like epithelial cornification, perhaps indicating apoptosis of keratinocytes or other specific host cell population.
Putative model of COVID-19 pathogenesis
The network also suggests that the SARS-CoV-2 infection may trigger a lipopolysaccharide-mediated response that causes a rise in cytokine and chemokine levels. This leads to an intracellular influx of calcium that results in epithelial migration, cornification and apoptosis.
Chemokine abnormalities may thus be responsible for severe COVID-19 features, such as the mucosal immune response of the airways, inflammatory bowel disease, interstitial pneumonia and fibrosis, and eosinophilic pneumonia. The results are progressive dyspnea with acute respiratory failure in some cases, weight loss and other symptoms like fever, fatigue and night sweats.
What are the implications?
The dual RNA-seq analysis pipeline (dRAP) set up by the investigators was used to examine all the transcripts found in the sample, allowing simultaneous quantification of both host and viral transcriptomes, probably for the first time ever.
This new outlook revealed that the most strongly and consistently expressed transcripts were those that play essential roles in viral survival and propagation.”
These include the viral spike, membrane and nucleocapsid genes, required for viral attachment, cell entry, replication, assembly and release.
The most robust viral responses were found in BALF samples, rather than in lungs or PBMCs. The human responses in SARS-CoV-2 infection were found in BALF, again, rather than the other tissues. Among cell lines, the NHBE had the highest expression of viral transcripts, as well as a DEG profile closest to that of BALF.
This may indicate that this cell line may offer an acceptable proxy for human tissues. Additionally, BALF would appear to be the tissue of choice for gene expression quantification.
The coexpression analysis enabled by dRAP produced a coherent hypothesis that predicted a possible mechanism by which COVID-19 may progress from the initial SARS-CoV-2 infection to patient symptoms. Taken together, these findings shed light on the molecular pathways implicated in COVID-19.”
This study thus also demonstrates the importance of this method for exploring multiple transcriptomes simultaneously.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources