A team of researchers at Nanjing University in China used several assays, atomic force microscopy, and simulations to determine that the N501Y mutation in the virus’s receptor-binding domain (RBD) increased interaction of the virus with the angiotensin-converting enzyme 2 (ACE2) receptor.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the ongoing coronavirus disease 2019 (COVID-19) pandemic, is an enveloped RNA virus. The spike protein, which has an amino-terminal subunit S1 and a carboxyl-terminal subunit S2, on the envelope, helps the virus infect host cells.
The S1 subunit on the RBD recognizes and binds to the host cell and is key in the zoonotic transmission of coronaviruses and infectivity of humans. The S2 subunit is responsible for membrane fusion. The first step in infection by SARS-CoV-2 is the RBD interaction with the human ACE2 cell receptor.
Two new variants of the virus, which apparently have increased transmissibility, have been recently discovered in the United Kingdom (B.1.1.7) and South Africa (B.1.351). Both have the N501Y mutation on the spike RBD, and the South African mutant has two more mutations, K417N and E484K, on the RBD.
Reports indicate that the approved mRNA vaccines have a high level of neutralization of the B.1.1.7 variant, but a lower level for the B.1.351 variant. Studies on neutralizing antibodies in Phase II and Phase III trials indicate they are not able to neutralize B.1.351. Because the mutations are on the RBD, it is important to understand how exactly they bind to ACE2.
Binding affinity of mutations to ACE2
The researchers used cell binding assays, kinetics, and simulations to understand the binding of the mutant viruses to ACE2. They reported their results in a paper published on the bioRxiv* preprint server.
Confocal microscopy of ACE2 in HEK293 cells showed ACE2 is mainly present in the membrane and endoplasmic reticulum. Cell surface-binding assays using fluorescent labeling clearly showed the binding of ACE2 to the RBD.
Using competition binding assay, the team found that the N501Y in the B.1.1.7 mutant showed a 4-fold higher binding affinity to ACE2 compared to the wild-type virus RBD. Mutations K417N and E484K showed a slightly lower binding affinity. When all the three mutations were present together, the binding affinity was similar to that of the wild type.
The authors used atomic force microscopy-based single-molecule force spectroscopy (AFM-SMFS) to measure the binding strength of the RBDs with the three mutations to ACE2. The RBD is modified to allow attachment to a peptide-coated AFM tip. The ACE2 expressing HEK293 cells were bound on a petri dish. When the AFM tip coated with RBD is moved toward the petri dish, a binding between ACE2 and RBD occurs. When the tip moves back, the interactions are broken. The team obtained an unbinding force map by doing this hundreds of times on the surface.
They found that the RBD mutants with N501Y and all three mutations had higher unbinding force than that of the RBD without the mutant. Calculations showed that the bond dissociation rates of both types of mutant RBDs are slower than that of the wild-type RBD. Surface plasmon resonance assay revealed all the RBD mutations had a higher binding affinity, an almost 10-fold increase, to ACE2 than the wild-type virus.
The researchers also built models for the RBD mutants and ACE2 complex and used steered molecular dynamics (SMD) simulations to investigate unbinding mechanisms under force. Upon pulling the RBD with the N501Y mutation, after a certain force, it changed its conformation and formed another interaction with Y501 and Y41 of ACE2. The N501Y may form a p-p interaction with Y41 of ACE2, leading to a stronger interaction.
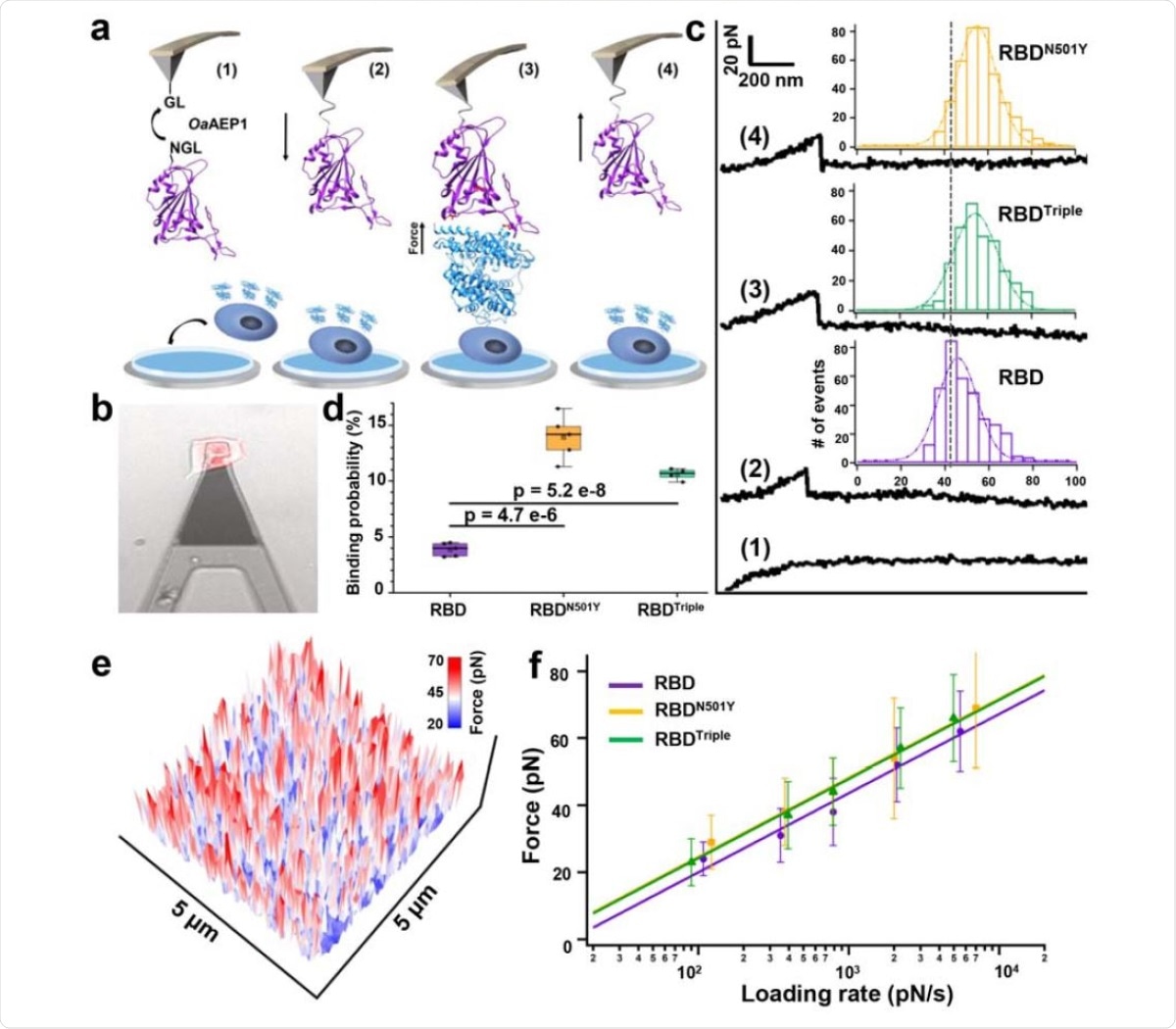
AFM-SMFS experiment to quantify the strength between RBDs and ACE2 on the living cell. a) Schematics of AFM-SMFS measurement processes show how the interaction is quantified. RBD with an N-terminal NGL is immobilized on a GL-coated AFM tip by ligase OaAEP1, which recognizes the two sequences and ligates them into a peptide bond 1). By approaching the AFM tip to the target cell 2), RBD binds to ACE2 3). Then the tip retracts, and the complex dissociates finally, leading to an unbinding force peak 4). b) Image showed the reddish ACE2-mCherry transfected HEK293 cell being measured under the tip by an inverted fluorescent microscope. c) Representative force-extension curves show no binding event (curve 1) and specific binding events between RBD-ACE2 complexes with an unbinding force peak (curves 2-4). From the force histogram (inset), RBDN501Y and RBDTriple showed higher forces as 57 pN and 56 pN than the RDB (49 pN). d) Box plot of the specific binding probabilities between the three RBDs and cell indicated a higher probability for the two mutants from AFM experiments under all five different velocities. The box indicates the 25th and 75th percentiles. e) f) The plot between loading rate and most probable unbinding forces from the complexes showed a linear relationship. The data is fitted to the Bell-Evans model to extract the off-rate.
Mutations increase virus binding to ACE2
Thus, all the different tests show that the N501Y is more important than the other two mutations in binding to ACE2. While K417N decreased the interaction, E484K slightly increased the interaction. Thus, when all three mutations are present, the net effect is similar to that when only the N501Y mutation is present.
In another virus variant originating in Brazil identified in January 2021, the N501Y mutation is present, apart from other mutations.
Consequently, we believe that the N501Y is a critical mutation to affect the transmission of COVID-19 by strengthening the interaction between RBD and ACE2,” write the authors.
This mutation will help the virus bind to the host for longer, allowing it more time to fuse with the host cell membrane, thus increasing infectivity.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Tian, F. et al. (2021) Mutation N501Y in RBD of Spike Protein Strengthens the Interaction between COVID-19 and its Receptor ACE2. bioRxiv. https://doi.org/10.1101/2021.02.14.431117, https://www.biorxiv.org/content/10.1101/2021.02.14.431117v1
- Peer reviewed and published scientific report.
Tian, Fang, Bei Tong, Liang Sun, Shengchao Shi, Bin Zheng, Zibin Wang, Xianchi Dong, and Peng Zheng. 2021. “N501Y Mutation of Spike Protein in SARS-CoV-2 Strengthens Its Binding to Receptor ACE2.” ELife 10 (August): e69091. https://doi.org/10.7554/eLife.69091. https://elifesciences.org/articles/69091.