The ongoing pandemic of coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has largely defied attempts to contain its spread by non-pharmaceutical interventions (NPIs). With the massive loss of life and economic damage, the only way out, in the absence of specific antiviral therapeutics, has been the development of vaccines to achieve population immunity.

 *Important notice: Preprints publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Preprints publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
The furin cleavage site
The SARS-CoV-2 is a betacoronavirus, and is most closely related to the bat SARS-related coronavirus (SARSr-CoV) represented by the genome sequence RaTG13, which shares 96% identity with the former. This has made the bat virus the most probable precursor of the virus in current circulation.
The origin of this strain is linked to the emergence of the novel furin cleavage site in the viral spike glycoprotein. The furin is a serine protease widely expressed in human cells, that cleaves the SARS-CoV-2 spike at the interface of its two subunits. It is encoded by a gene on chromosome 15.
Furin acts on substrates with single or paired basic residues during the processing of proteins within cells. Such a polybasic furin cleavage site is found in various proteins from many viruses, including Betacoronavirus Embecoviruses, and the Merbecovirus. However, within the betacoronaviruses of the sarbecovirus lineage B, this type of site is unique to SARS-CoV-2.
The study used a bioinformatic approach using the genomic data available on the National Center for Biotechnological Information (NCBI) databases, to identify the origin of the furin cleavage site.
Same ancestor
They found three coronaviruses that were very similar to the SARS-CoV-2 at the genomic level. These are Pangolin-CoVs (2017, 2019), Bat-SARS-like (CoVZC45, CoVZXC21) and bat RatG13.
The three genomic fingerprints used to identify these matches include fingerprint 1, in the orf1a RNA polymerase gene, including the nsp2 and nsp3 genes; fingerprint 2, at the beginning of S gene, covering the part encoding the N-terminal domain and the receptor-binding domain (RBD) that mediates attachment to the host cell receptor, the angiotensin-converting enzyme 2 (ACE2).; and fingerprint 3, the orf8 gene.
These fingerprints are distinctive to the three closely related coronaviruses only at the RNA level, but the amino acid sequences in the translated proteins are similar to other sarbecoviruses.
The sharing of these genomic sequences indicates their common ancestry, supported by other short sequence features, with one deletion and three insertions. All three strains show the same deletion-insertion pattern at the same four different locations in the spike gene.
Spike gene recombination in a common ancestor
The analysis of the phylogeny of these three strains showed that the first to diverge was the pangolin coronavirus, with the RatG13 being the closest. However, when only the spike is analyzed, there is a high similarity between the pangolin CoV, RaTG13 and SARS-CoV-2.
This may indicate the occurrence of recombination events between the Pangolin-CoV (2017) and RatG13 ancestors. This was followed by the shift of the pangolin CoV to pangolin hosts.
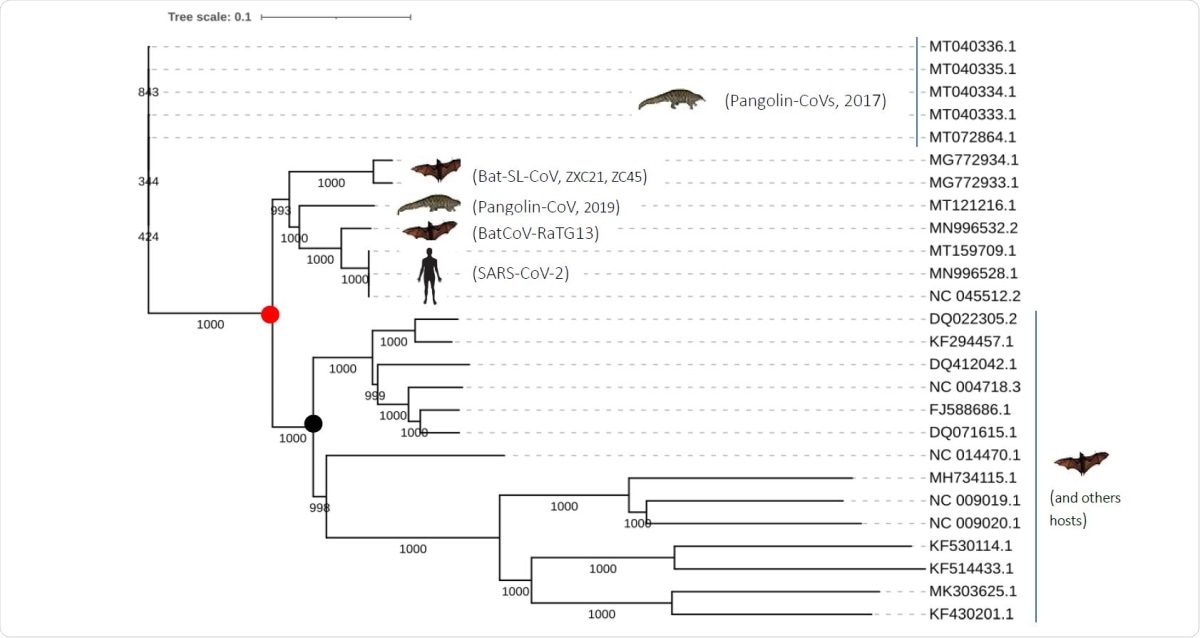
Phylogenetic tree of the closely related SARS-CoV-2 coronaviruses based on complete genomes.
Unique codons encoding arginines in the furin cleavage site
The furin cleavage site consists of four amino acids PRRA, which are encoded by 12 inserted nucleotides in the S gene. A characteristic feature of this site is an arginine doublet.
This insertion could have occurred by random insertion mutation, recombination or by laboratory insertion. The researchers say the possibility of random insertion is too low to explain the origin of this motif.
Surprisingly, the CGGCGG codons encoding the two arginines of the doublet in SARS-CoV-2 are not found in any of the furin sites in other viral proteins expressed by a wide range of viruses.
Even within the SARS-CoV-2, where arginine is encoded by six codons, only a minority of arginine residues are encoded by the CGG codon. Again, only two of the 42 arginines in the SARS-CoV-2 spike are encoded by this codon – and these are in the PRRA motif.
For recombination to occur, there must be a donor, from another furin site and probably from another virus. In the absence of a known virus containing this arginine doublet encoded by the CGGCGG codons, the researchers discount the recombination theory as the mechanism underlying the emergence of PRRA in SARS-CoV-2.
Time of acquisition
The second question is when this shift occurred. The RaTG13 virus was isolated in 2013, which could indicate that this site was acquired after that, giving rise to the current SARS-CoV-2 strain. This could mean it occurred within the bat host before it leaped the species barrier, or within the human host itself.
The first scenario is supported by the finding of the same RBD modifications in bat and human viruses, with three O-linked glycans around the furin site having been acquired in both. Both viruses also show completely identical sequences around this site.
To add weight to this hypothesis, however, a crucial piece of evidence is missing. RaTG13 sequences acquired in 2021 need to be analyzed to find whether the furin site was acquired by this virus, as well as the SARS-CoV-2 ancestor in the bat.
Conclusion
Describing this mystery site as “a furin site that has changed the world,” the researchers sum up:
All these lines of evidence and reasoning show that the acquisition of the polybasic furin cleavage site by SARS-CoV-2 is a “missing link” in our understanding of its evolutionary history, that can only be addressed through the discovery of new viruses.”

 *Important notice: Preprints publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Preprints publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.