The rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected over 111 million individuals worldwide and has resulted in the COVID-19 pandemic. Many pangolin and bat-derived viruses are closely related to SARS-CoV-2, which suggests its likely zoonotic origin.
Researchers have found a 96% similarity in nucleotide sequence between bat coronavirus (RaTG13), isolated from

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
s in China in 2013, and SARS-CoV-2 and ~97% amino acid similarity with the Spike (S) protein. This S protein performs two essential roles, i.e., mediates receptor-binding and membrane fusion. Hence, spike protein is regarded as a key coronavirus determinant of host tropism.
ddd
Similarly, a high percentage of similarity was also found in viruses present in Manis javanica (pangolins). A 97.4% amino acid concordance has been reported to be present in the receptor-binding domain (RBD) of the spike protein found in the pangolin virus.
A study of various epidemics over a couple of decades, i.e., the emergence of swine acute diarrhea syndrome coronavirus (SADS-CoV), severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV) have shown the world the epidemic potential of coronaviruses.
Scientists have explained that a minor change in the virus's genetic sequence may play a crucial role in its adaptation to a new host. A minor modification in only two amino acids established a necessary change in SARS-CoV and MERS-CoV spike proteins required to adapt to human beings.
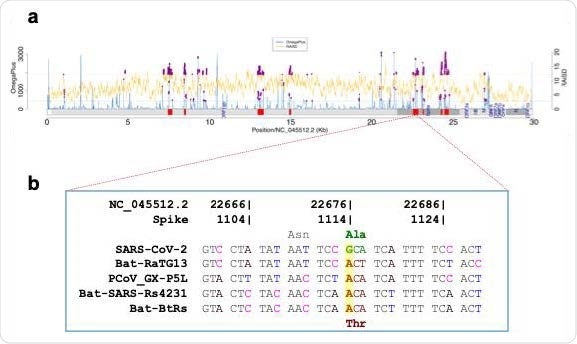
Fig 1. Selective sweeps analysis. (a) Selective sweep regions (shown as red blocks) identified in 182,792 SARSSARS-CoVCoV-2 genomes, using OmegaPlus (blue lines) and RAiSD (yellow lines). The common outliers (0.05 cutoff, purple dots) from the two methods were used to define selective sweep regions. (b) Non-synonymous difference (Thr372Ala) between SARS SARS-CoVCoV-2 and four other Sarbecovirus members found in the putative selective sweep region (22,529 22,529-22,862).
A similar change was also found in many other viruses, such as Ebola, where a single alanine-to-valine mutation at position 82 in the glycoprotein initiated their human adaptation, which caused the outbreak. Many recent outbreaks associated with an RNA virus, for example, West Nile virus, Zika virus, and Chikungunya virus, also occurred due to a single amino acid change in the virus nucleotide sequence.
In the case of SARS-CoV-2, a single mutation at position 614 in the S protein caused a change in the production of single aspartic acid to glycine, which has resulted in an increase of replication vigor in humans. However, the genetic determinants of SARS-CoV-2 responsible for the change of its host from animal to human remained unknown.
Scientists believe a strong signature of positive selection occurs when a virus changes its host through rapid evolution or cross-species transmission.
Such incidence was reported for the brief SARS epidemic that took place in 2002–2003. During this period, a series of adaptive changes or mutations occurred in SARS-CoV genomes characterized based on dN/dS tests.
These tests are designed for the comparison of eukaryotic interspecies. However, the limitation of these tests is that they cannot detect the hallmark signature of positive selection in viral lineages with small sequence divergence.
Recently, researchers have used highly sophisticated methods that are capable of detecting selective sweeps. In this method, a selectively favorable mutation spreads through the population, causing a reduction in the level of sequence variability at nearby genomic sites.
With the help of unprecedented statistical tools, scientists have analyzed more than 182,000 SARS-CoV-2 genomes and found that positive selection plays a vital role in the adaptive evolution of SARS-CoV-2. Considering the coronavirus host tropism, researchers provided strong evidence that the spike protein residue 372 contributes to adaptive mutation, which in turn aggregates their replication in human lung cells. The research by scientists in the U.S and Israel is posted online on the bioRxiv* preprint server.
The genetic modification, i.e., threonine-to-alanine change, may enable the virus to replicate more vigorously within human cells. This also promotes an efficient human-to-human transmission.
Prior computation-based research and the study of pseudotyped viruses have identified positive selection in SARS-CoV-2.
Even though a study of the pseudoviruses provided vast volumes of useful information, the entire life cycle of the virus or the interactions between different viral proteins and the host were not obtained. Recently, the use of live viruses has helped the development of hamster models that recapitulate human-to-human transmission.
In summary, current research shows the presence of a distinct footprint of positive selection around a non-synonymous change (A1114G; T372A) within the RBD of the SARS-CoV-2 spike protein. This plays a vital role in incapacitating species barriers and also achieving interspecies transmission from animals to humans.
Further, the structural analysis also indicates that the change of threonine with alanine in SARS-CoV-2 removes the glycosylation site at N370. Such changes indicate a favorable binding of the virus to the cellular receptor in humans (ACE2).
Another interesting suggestion was that, unlike previous assumptions, the Huanan seafood market, China, might not be the origin point of the novel SARS-CoV-2. The transmission may have occurred unnoticed elsewhere for a period that provided the ancestor virus enough time for their adaptation to human replications.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
A selective sweep in the Spike gene has driven SARS-CoV-2 human adaptation Lin Kang, Guijuan He, Amanda K. Sharp, Xiaofeng Wang, Anne M. Brown, Pawel Michalak, James Weger-Lucarelli bioRxiv 2021.02.13.431090; doi: https://doi.org/10.1101/2021.02.13.431090, https://www.biorxiv.org/content/10.1101/2021.02.13.431090v1
- Peer reviewed and published scientific report.
Kang, Lin, Guijuan He, Amanda K. Sharp, Xiaofeng Wang, Anne M. Brown, Pawel Michalak, and James Weger-Lucarelli. 2021. “A Selective Sweep in the Spike Gene Has Driven SARS-CoV-2 Human Adaptation.” Cell 184 (17): 4392-4400.e4. https://doi.org/10.1016/j.cell.2021.07.007. https://www.cell.com/cell/fulltext/S0092-8674(21)00833-3?.