Some of the genes in the human genome appear to confer a protective effect against severe coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). A new paper published online in the journal PNAS describes genes involved in innate immunity that have undergone variations between different human subgroups.
Infectious diseases vary in prevalence with environmental conditions, which vary dramatically in different parts of the world. These act as selective factors on gene variants, such as the toll-like receptor gene variants that reduce the susceptibility to some bacterial infections and allergies.
Again, the host cell proteins that interact with RNA viruses are encoded in various regions of the human DNA, and as such, these viruses might have stimulated many such changes.
COVID-19 and chromosome 3 haplotype
An earlier study showed an association between a haplotype on chromosome 3 and risk of critical COVID-19 illness. The risk of intensive care unit (ICU) admission following SARS-CoV-2 was doubled with each copy of this variant.
This gene was found to be present in 65% of carriers in South Asia but 16% in Europe, but remarkably, it is absent in East Asia. The researchers hypothesize that this is due to a beneficial effect of the haplotype in South Asia against other earlier pathogens, but was now negatively selected in East Asia.
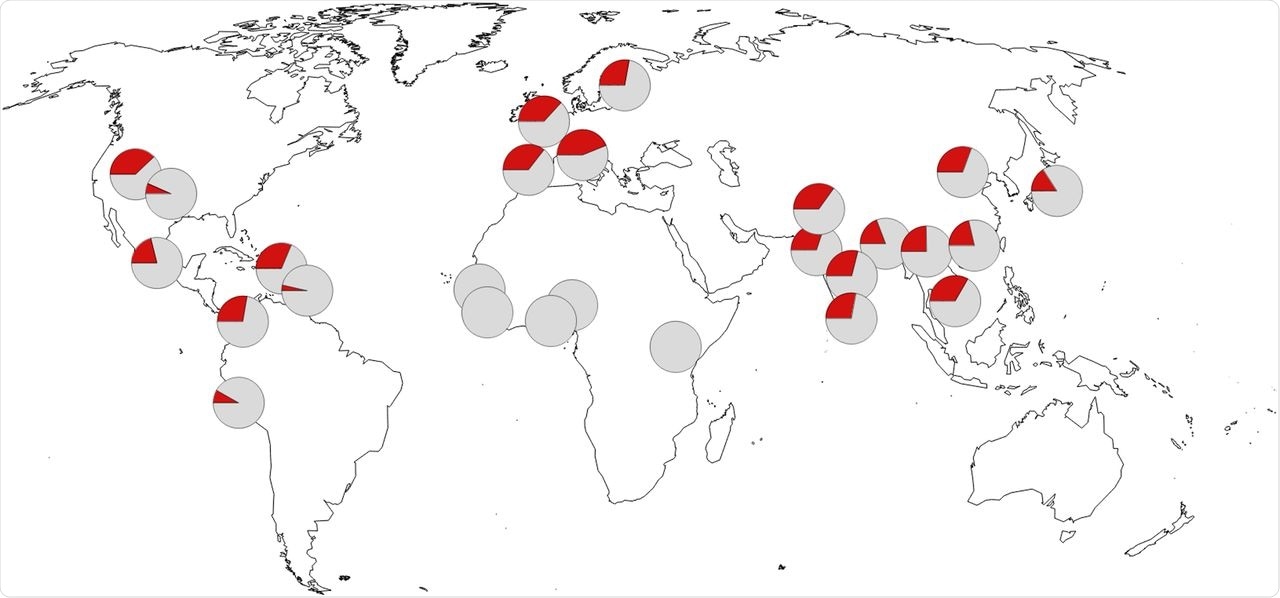
Geographic distribution of the allele indicative of the Neandertal haplotype protective against severe COVID-19. Pie charts indicate minor allele frequency in red at rs1156361. Frequency data are from the 1000 Genomes Project (23). Map source data are from OpenStreetMap.
Neanderthal SNPs confer protection
The current study, from the Genetic of Mortality in Critical Care (GenOMICC) consortium, examined over 2,200 critical COVID-19 patients and controls. This study showed a risk region on chromosome 3, with seven loci that affect the whole genome, located on chromosomes 6, 12, 19, and 21.
The mutations at loci on chromosomes 6, 19, and 21 are single-nucleotide polymorphisms (SNPs) that increase the risk of ICU admissions in COVID-19 patients and match alleles found in human DNA thought to be of Neanderthal origin. Yet, these are not found in the genomes of over a hundred African Yoruba people as available on the 1000 Genomes Project.
Chromosome 12 SNPs came from Neanderthals
The researchers compared this with data from the COVID-19 Host Genetics Initiative (HGI). This showed that SNPs on chromosome 12 that were linked to the hospitalization in COVID-19 patients were in linkage disequilibrium in Europeans. This 75 kb haplotype was compared to present-day and older genomes.
The study used haplotypes found ten or more times among individuals in the 1000 Genomes Project and among genomes from Neanderthal and Denisovan skeletons from caves in Siberia and Croatia. Using phylogenetic techniques, they found that eight of 64 modern human haplotypes were closely related to the three older skeletons.
The researchers then carried out a calculation meant to distinguish a segment of the genome that came from a common ancestor of modern humans and Neanderthals, from one which was incorporated into the modern human genome from Neanderthals by intermixing of the groups. They found that a segment of 75 kb would probably not have survived that long if it had descended from a common ancestor.
“We thus conclude that the haplotype entered the human gene pool from Neandertals.”
Neanderthal variant reduces ICU risk in COVID-19
The study also found that the index variant of the protective haplotype was similar to that found in all three Neanderthal genomes and reduced the risk of ICU admission by over a fifth with each copy. The Neanderthal genomes contained 34 of 35 SNPs that cosegregate with the protective SNP variant.
This haplotype is found in 25% to 30% of most Eurasian populations but is almost zero in sub-Saharan Africa. It is present, though at lower levels, in some populations ethnically related to Africa. This could be due to gene inflows from European or Native American populations, in this case.
Possible mechanism of protection
The protective haplotype contains parts of three genes, called OAS1, OAS2, and OAS3, that encode enzymes induced by interferons and activated by double-stranded RNAs. The effect of these enzymes, called oligoadenylate synthetases, is to produce oligoadenyl chains that trigger ribonuclease L, breaking down double-stranded RNA within the cell, a classical antiviral mechanism in infected cells. Other antiviral pathways are also activated by ribonuclease L.
Some Neanderthal alleles appear to be functionally important, such as an SNP that affects a splice affector site in OAS1. This has been earlier found to be protective against West Nile virus. The replacement of the ancestral allele present in the Neanderthal haplotype with other variants leads to less protective isoforms of this protein.
This ancestral allele is currently present only in this context outside Africa. Interestingly, in Africans today, the ancestral allele is found only as a genetic variant independent of the Neanderthal haplotype.
The Neanderthal haplotype also appears to confer more resistance to hepatitis C. Again, the OAS1 missense variant is protective against SARS-CoV, according to a small study.
Temporal patterns in haplotype frequencies
Using thousands of genomes from earlier human fossils, the researchers claim that the Neanderthal OAS haplotype has increased in frequency from 10% to 30% over the centuries, with the last increase in Eurasian populations appearing to have occurred quite recently.
More study will be required, using data from fossils found other than western Eurasia and tracing the allele frequencies over time, as well as adjusting for migrations of large population groups that account for changes in allele frequencies over entire genomes.
What are the implications?
The chromosome 12 Neanderthal haplotype is thus protective against severe COVID-19 and is found at frequencies of 50% or more in Eurasia and the Americas. The increased enzymatic activity of the ancestral variants may have been benefited the later populations that acquired this through genetic mixing, as they were exposed to new RNA viruses.
It is interesting that the Neandertal-derived OAS locus has recently increased in frequency in Eurasia... suggesting that this locus has been positively selected.”
Further study will confirm or refute this hypothesis.