A team of scientists from the USA and Germany has recently studied the evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a representative set of sequences from the USA collected between 2020 and 2021. The findings reveal that the viral genome has accumulated multiple mutations over time with only occasional loss of mutation. The main driving forces behind such genetic variations include widespread infection and superspreader events. The study is currently available on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Within one year of its emergence, SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), has infected 110 million people and claimed 2.4 million lives globally. SARS-CoV-2 is a single-stranded, positive-sense, enveloped virus of the Coronaviridae family. Among various proteins present on the viral envelope, the spike glycoprotein is the most immunogenic component because of its direct involvement in the viral recognition and entry processes. Any alteration in the amino acid sequences of the viral open reading frames (ORFs), which encode essential viral proteins, can lead to the development of new viral variants. Compared to other RNA viruses, mutations occur less frequently in SARS-CoV-2 because of the presence of 3’-5’ exoribonuclease proofreading ability. However, evidence suggests that most of the single nucleotide substitutions observed in SARS-CoV-2 are likely caused by RNA editing deaminases, which generally target adenine and cytosine bases to cause transition mutations. In addition, several recombination mutations via template strand switching have been documented in SARS-CoV-2.
In the current study, the scientists have investigated the incidence of SARS-CoV-2 mutations that appeared during 2020 in the United States and derived a set of mutational signatures representing distinct viral variants. Based on the mutational signatures, they have aimed to identify new variants or new mutations in previous variants that have been introduced from different regions worldwide. They have studied a representative set of sequences that cover the entire SARS-CoV-2 genome in the United States.
Important observations
For the analysis, the scientists collected more than 8000 full-length SARS-CoV-2 sequences from COVID-19 patients between January 2020 and January 2021. They identified multiple distinct SARS-CoV-2 variants, including the original Wuhan strain and its subvariants carrying minor mutations; and two varieties of the European strain with D614G mutation. The European strain rapidly acquired multiple mutations that ultimately resulted in a new homegrown dominant variant, s48. They observed that instead of a recombination event, these mutations actually resulted from the acquisition of single nucleotide substitutions that rapidly increased in frequency due to superspreader events.
Importantly, they observed that the major USA variants accumulated an increasing number of mutations over time, indicating the fact that the emergence of novel mutations can increase with uncontrolled viral transmission and that the newly emerged variants may influence the effectiveness of therapeutics antibodies and vaccines. Specifically, they observed that during 2020, more than 20 amino acid substitution mutations occurred in the spike protein, and many of these mutations are still remaining in the population at a low frequency. This indicates that these substitution mutations are increasingly accumulating over time with a minimal loss from the population through genetic drift.
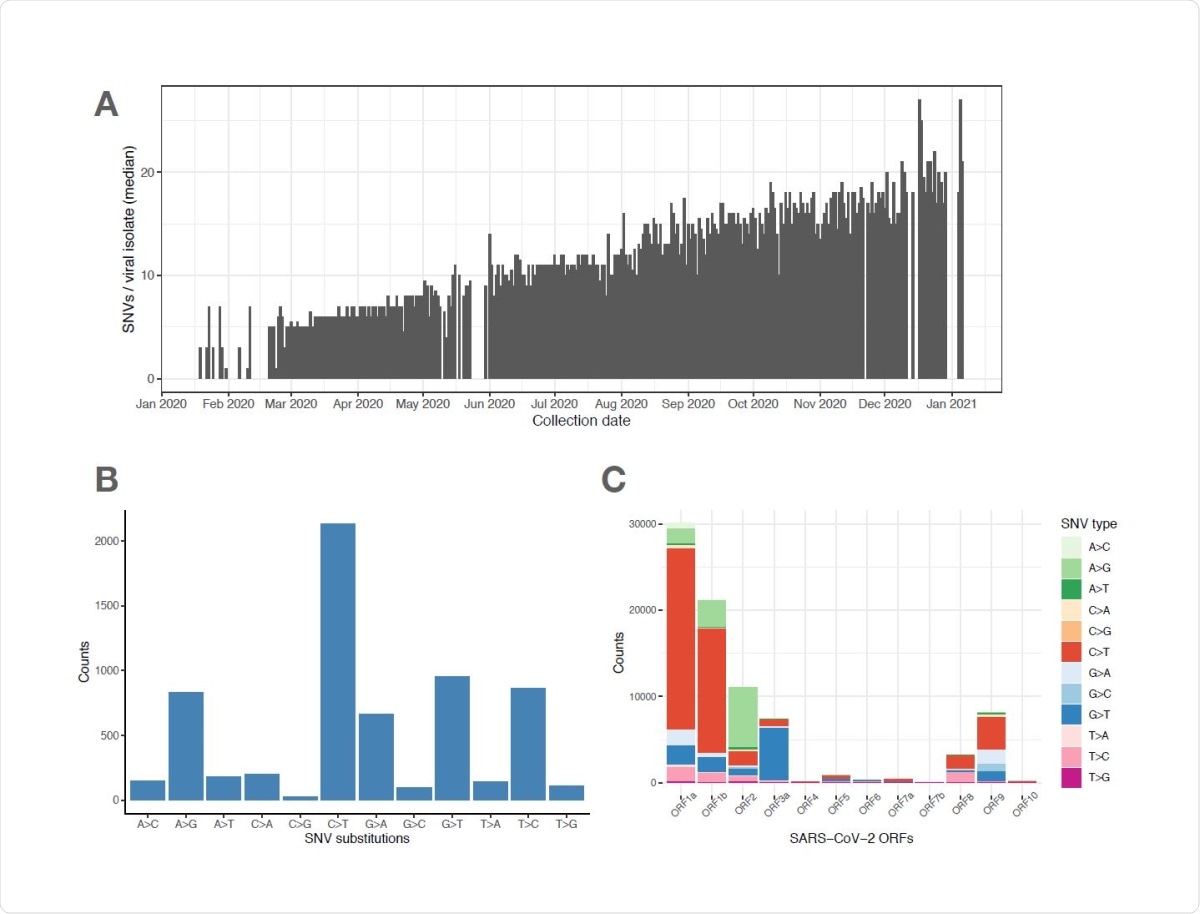
SARS-CoV-2 viral genomes accumulate specific sets of SNVs over time. (A) Frequency histogram showing the steady increase of SNVs called per viral isolate over time (Collection Date), indicating their aggregation in SARS-CoV-2 genomes. (B) Distribution of substitutions at unique SNVs. Two of the most frequent SNV substitutions, C>T and A>G, have been previously associated with APOBEC and ADAR deaminase activities, on the SARS-CoV733 2 ssRNA(+) genome or its dsRNA intermediate, respectively. (C) Graphical representation of SNV substitution profiles at various SARS-CoV-2 ORFs, illustrating intrinsic mutational bias for C>T dominating the mutation pattern in some ORF’s (i.e. 1a and 1b), but being masked (likely by selection) in other ORF’s like ORF2 encoding Spike region.
The scientists mentioned that these low frequency spike variants could eventually become the dominant variants because of the increasing number of superspreader events in the USA. Upon achieving a high-frequency level due to superspreader events, these variants can undergo positive selection in viral evolution, leading to the emergence of variants capable of escaping the host immune responses. Such variants could appear in people receiving inadequate vaccine doses.
Among distinct spike variants, the scientists identified a novel B1.375 variant containing the H69/V70 deletion mutation. They considered B1.375 as a variant of concern because H69/V70 deletion may increase the possibility of acquisition of new mutations with clinical relevance. Moreover, they identified certain non-spike variants containing mutations in ORF1a and 1b, potentially influencing the disease severity and viral transmissibility.
To established distinct mutational signatures, they analyzed a number of silent mutations that change only the DNA or RNA sequence without altering the amino acid residue (synonymous mutations). Their analysis revealed that the most frequent mutations were C-to-U and U-to-C transitions. They believe that RNA modification events by RNA editing enzymes are primarily responsible for these mutations.
Study significance
The study highlights that superspreader events are potentially responsible for generating high-frequency novel spike variants from the pre-existing reservoir of low-frequency variants. Therefore, strict measures should be taken to avoid such events.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Rafail Nikolaos Tasakis, Georgios Samaras, Anna Jamison, Michelle Lee, Alexandra Paulus, Gabrielle Whitehouse, Laurent Verkoczy, F. Nina Papavasiliou, Marilyn Diaz. 2021. SARS-CoV-2 variant evolution in the United States: High accumulation of viral mutations over time likely through serial Founder Events and mutational bursts. BioRxiv. doi: https://doi.org/10.1101/2021.02.19.431311, https://www.biorxiv.org/content/10.1101/2021.02.19.431311v1
- Peer reviewed and published scientific report.
Tasakis, Rafail Nikolaos, Georgios Samaras, Anna Jamison, Michelle Lee, Alexandra Paulus, Gabrielle Whitehouse, Laurent Verkoczy, F. Nina Papavasiliou, and Marilyn Diaz. 2021. “SARS-CoV-2 Variant Evolution in the United States: High Accumulation of Viral Mutations over Time Likely through Serial Founder Events and Mutational Bursts.” Edited by Yury E. Khudyakov. PLOS ONE 16 (7): e0255169. https://doi.org/10.1371/journal.pone.0255169. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0255169.