A team of scientists from the United States recently investigated the mutational signatures in the receptor-binding domain (RBD) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein. The findings reveal that the frequency of L452R mutation in the spike region has increased since November 2020. The L452R mutation, which is predicted to increase viral infectivity and host immune evasion potency, has been found to associate with a viral variant that has recently caused massive outbreaks in California. The study is currently available on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), is an enveloped RNA virus of the Coronaviridae family. Apart from the original Wuhan strain that emerged in December 2019, many distinct variants of SARS-CoV-2 containing several spike mutations are continuously appearing throughout the course of the pandemic. The majority of these variants contain mutations in the RBD of the spike protein, and thus, are considered as “variants of concern” because of their ability to increase viral infectivity, virulence, and immune evasion potency.
Of two domains (S1 and S2) of SARS-CoV-2 spike glycoprotein, S1 interacts with the host cell angiotensin-converting enzyme 2 (ACE2) receptor via the RBD. This is followed by proteolytic dissociation of S1 and S2 domains and subsequent fusion of the viral envelope with the host cell membrane. Because the RBD is significantly involved in viral recognition and entry, any structural changes in this region due to mutation can influence viral transmissibility and virulence. Moreover, antibodies developed against the RBD have been found to have maximum potency in neutralizing SARS-CoV-2.
In the current study, the scientists have analyzed the genetic alterations in the 414-583 amino acid region of the spike S1 domain, which partially covers the RBD. Using polymerase chain reaction (PCR) and Sanger sequencing methods, they have analyzed a total of 570 nasopharyngeal specimens collected from COVID-19 patients between April 2020 and February 2021, from different regions of the United States.
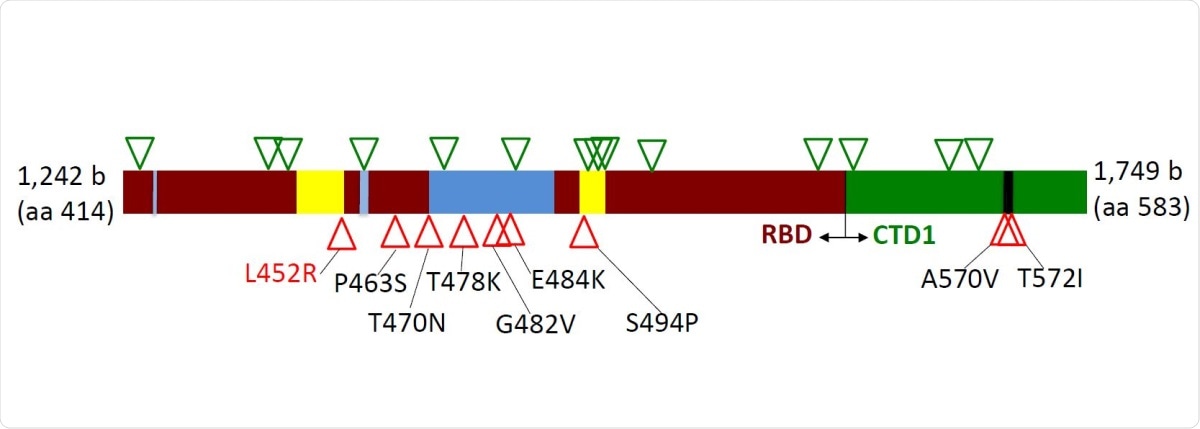
Distribution of silent (green triangles) and amino acid (red triangles) mutations across region 414-583 of the Spike protein. Dark red – receptor-binding domain (RBD); Green – C-terminal domain 1 (CTD1) of S1 Spike region; Blue – receptor-binding ridge epitope residues; Yellow – 443-450 loop epitope residues; Black – 570-572 loop residues.
Important observations
The scientists identified L452R amino acid substitution in the spike region as the dominant mutation in specimens collected since November 2020. Specifically, they observed that two independent SARS-CoV-2 variants (CAL.20C and CAL.20A) containing spike L452R mutation emerged recently in the state of California. Of these variants, CAL.20C (clade 20C; lineage B.1.429) is considered to be the predominant variant in California since November 2020. However, the CAL.20A variant (clade 20A; lineage B.1.232) identified in this study has emerged much more recently than CAL.20C and is primarily circulating in California. Based on the phylogenetic analysis, the scientists indicated that L452R mutation is the primary driving force behind the emergence of both variants. Such an increase in L452R mutation frequency in recent SARS-CoV-2 variants directly indicates its crucial involvement in viral adaptive evolution due to positive selection.
In contrast to CAL.20C, no massive clonal expansion was observed for CAL.20A. According to the study findings, CAL.20C contains two additional spike mutations along with L452R, which are missing in CAL.20A. The scientists believe that these additional mutations may be responsible for increasing the adaptive benefits of L452R, and because of this reason, CAL.20A could not achieve the same expansion rate as CAL.20C.
The scientists examined the Nextstrain and GISAID databases and found that apart from two California variants, L452R mutation is present in more than 400 SARS-CoV-2 genomes isolated from over 20 countries. This indicates a strong positive selection for L452R mutation. Interestingly, they found CAL.20A variant from a gorilla at the San Diego Zoo, which contains two additional mutations in the non-structural protein 2 (NSP2).
Based on the available literature, the scientists mentioned that the substitution of hydrophobic leucine residue at position 452 with a polar, highly hydrophilic arginine residue (L452R mutation) might strongly favor the interaction between virus and host cell, which in turn may significantly increase the viral transmissibility and infectivity. Moreover, such amino acid substitution is expected to reduce the virus-neutralizing ability of antibodies specifically targeting the spike RBD.
Study significance
The study demonstrates the importance of L452R mutation in viral adaptive evolution. A strong positive selection for L452R mutation has been particularly observed only recently, indicating that the viral adaptive evolution might have occurred in response to extensive containment measures or increasing population-level immunity against the original viral strains.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources