As the COVID-19 pandemic continues to spread worldwide, several new mutations of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have emerged. Of note are the B.1.1.7 lineage discovered in the United Kingdom (UK) and the B.1.351 that emerged in South Africa late in 2020. The B.1.1.7 variant has now spread to more than 50 countries, and both mutations are believed to increase the transmissibility of the virus.
Some studies have reported that neutralizing antibodies, particularly the neutralizing antibodies directed to the N-terminal domain, have a lower effect on these variants. However, many antibodies to the virus receptor-binding domain (RBD) seem to retain most of their potency.
In a previous study, researchers reported two extremely potent RBD antibodies, 1-57 and 2-7, whose effect was not inhibited by the new variants. To understand how these antibodies are effective against the new strains, they investigated their structures using cryo-electron microscopy (EM). They report their results in a research paper published on the bioRxiv* preprint server.
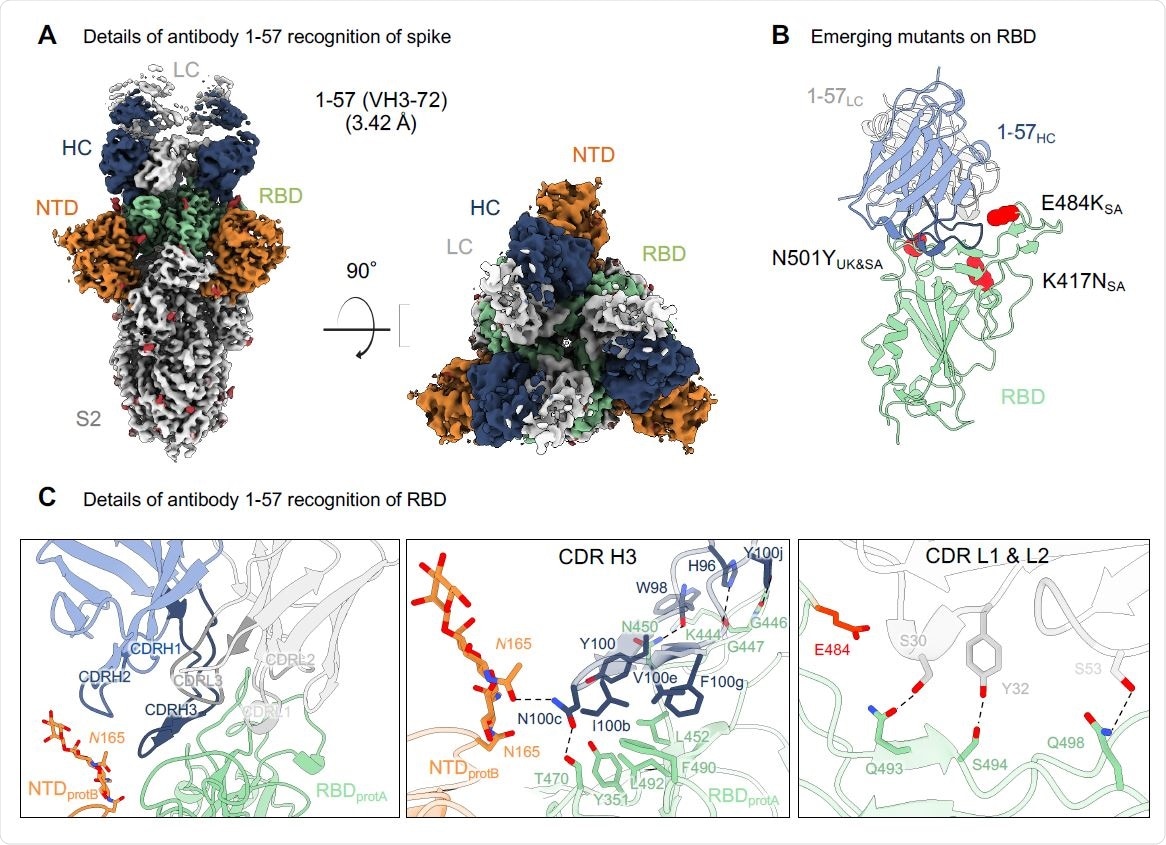
Antibody 1-57 utilizes a hydrophilic pocket to accommodate mutation E484K in emerging strains. (A) Cryo-EM reconstruction for spike complex with antibody 1-57 from two orthogonal views; a single conformation with all RBDs down is observed. NTD is shown in orange, RBD in green, glycans in red, antibody heavy chain in blue and light chain in gray. (B) Domain level view of 1-57 in complex with RBD, with the emerging mutants highlighted in red. (C) Details of antibody 1-57 recognition of RBD showing the overall interface (left panel), recognition by CDR H3 (middle panel) and recognition by CDR L1 and L2 (right panel). CDR H1, H2, H3 are colored in shades of blue; CDR L1, L2, and L3 are colored in shades of gray. E484 is highlighted in bright red (right panel). Nitrogen atoms are colored in blue, oxygen atoms in red; hydrogen bonds (distance <3.2 Å) are represented as dashed lines.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Understanding antibody binding complexes
Using SARS-CoV-2 spike protein complexes with antibody 1-57, the team reconstructed the structure at a resolution of 3.42 Å (roughly three ten-billionths of a meter). They identified a single conformation with three antigen-binding fragments per trimer. Each fragment was bound to an RBD in the down configuration.
The primary interaction of the RBD to the antibody was via the CDR H3, forming disulfide bonds, hydrophobic interactions, and hydrogen bonds. Of the mutations in the UK and South African variants, only E484K of the South African strain was close to the binding site of 1-57. Structural modeling revealed a lysine residue was geometrically compatible with 1-57 binding.
For the 2-7 antibody, cryoEM analysis revealed three fragments bound to a single spike, with two bound to the RBD in the up configuration and one to the RBD in the down configuration. The interactions with the RBD were predominantly with the residues 438-451 and 495-502. CDR H2 formed several salt bridges and hydrogen bonds. In the UK and South African mutations, antibodies 2-7 only bound near N501.
The antibodies 1-57 and 2-7 used heavy chain genes and were not frequent in the SARS-CoV-2 specific group of antibodies. This suggests antibodies that are genetically similar to 1-57 and 2-7 may not be common in the human response to the virus.
Parts of the virus that are frequently a target of antibodies may cause the virus to develop evasion mechanisms. Using previously reported structures of antibody-RBD complexes, the authors found that epitopes of the antibodies 1-57 and 2-7 are targeted less often. These epitopes are also on regions that are not important for binding to the angiotensin-converting enzyme (ACE2) or for neutralizing antibody escape.
Although the antibodies 1-57 and 2-7 were not affected by the RBD mutations on the UK and South African variants, other RBD mutations can affect them. For example, mutations at positions 452 and 494 may affect 1-57 binding and mutation at position 439 may affect 2-7 binding. However, more research needs to be done to understand this further.
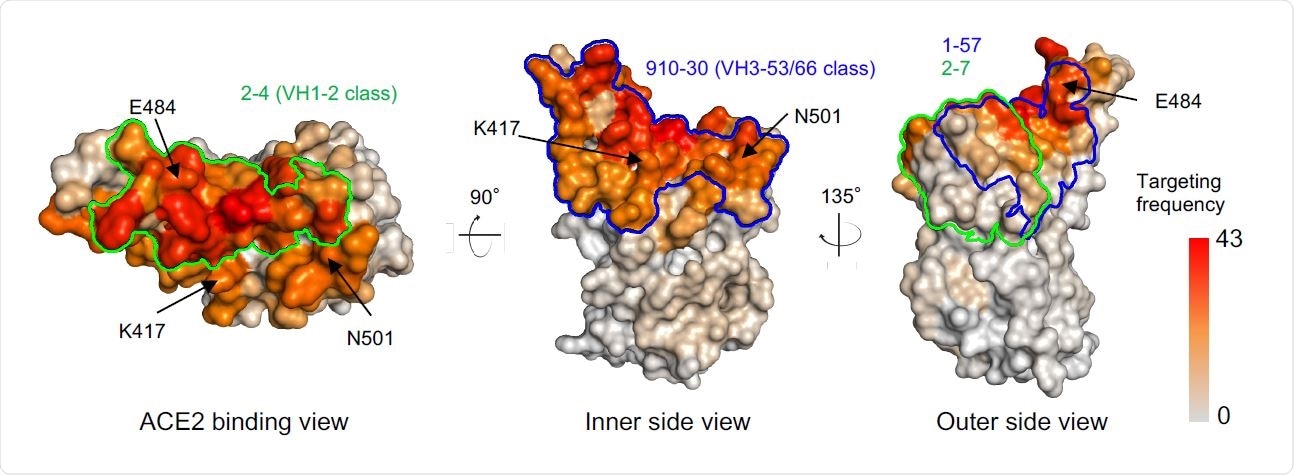
Comparison of 1-57 and 2-7 epitopes with antibody classes that exemplify high-frequency responses. Per residue frequency recognized by 52 known RBD-directed antibodies. VH1-2 and VH3-53/66 antibody classes recognize RBD residues with high targeting frequency. 1-57 and 2-7 recognize RBD residues with low targeting frequencies.
Some antibodies are effective against mutant strains
To understand if SARS-CoV-2 escape mutations are selected by neutralizing antibodies and antibody-resistant mutations are due to antibody selection, the team correlated the positional mutation frequency and antibody recognition frequency. They found only a weak correlation between the two, although there were higher mutation frequencies on the most frequently targeted residues.
The authors found a strong correlation between RBD mutations' frequencies and their effect on ACE2 binding affinity. This suggests RBD mutations may arise from selection advantage for ACE2 binding, helping the virus bind more strongly to ACE2.
However, since most RBD mutations are disadvantageous for ACE2 binding, the virus may have limited options for escaping neutralization by antibodies. This may explain the similar mutations in different SARS-CoV-2 lineages.
"Our analysis suggests that potent antibodies 2-7 and 1-57 may not put much selection pressure on circulating strains, thus resistance mutants to these two antibodies are less likely to arise," write the authors.
Since these two antibodies bind to virus regions, not on the ACE2-binding region, and their neutralizing capability is not affected much by mutations, they may be good candidates for developing therapeutics that can protect for a long time.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Cerutti, G. et al. (2021) Structural Basis for Accommodation of Emerging B.1.351 and B.1.1.7 Variants by Two Potent SARS-CoV-2 Neutralizing Antibodies. bioRxiv. https://doi.org/10.1101/2021.02.21.432168, https://www.biorxiv.org/content/10.1101/2021.02.21.432168v1
- Peer reviewed and published scientific report.
Cerutti, Gabriele, Micah Rapp, Yicheng Guo, Fabiana Bahna, Jude Bimela, Eswar R. Reddem, Jian Yu, et al. 2021. “Structural Basis for Accommodation of Emerging B.1.351 and B.1.1.7 Variants by Two Potent SARS-CoV-2 Neutralizing Antibodies.” Structure 29 (7): 655-663.e4. https://doi.org/10.1016/j.str.2021.05.014. https://www.cell.com/structure/fulltext/S0969-2126(21)00172-6.