Besides the highly publicized spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a number of other accessory proteins belonging to the virus are involved in pathogenesis by suppressing or evading the host immune response.
One such protein is open reading frame 7a (ORF7a), which has previously been implicated in interferon antagonism and may be involved in triggering the expression of pro-inflammatory cytokines.
A new study by researchers at Montana State University in the U.S., which was released as a preprint on the medRxiv* server, has identified a high variability in the C-terminal end of ORF7a in SARS-CoV-2 sequences collected from around the globe. They noted that mutations result in a defective suppression of interferon and lowered replication rate.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
These mutations were identified to have derived independently and not from a single lineage, suggesting that this mutation occurs innately. However, the mutation does not persist in the population, most likely due to the impaired suppression of the host immune response compared to wild-type virus.
How was the study performed?
The group gathered full SARS-CoV-2 genome sequences from 56 patients in Montana, USA, between April and July 2020 and composed a phylogenic tree using nearly 200,000 genome sequences from around the globe over the course of the pandemic for comparison.
Interestingly, only one of the 56 samples belonged to the original Wuhan lineage, with the remainder being of the B.1 lineage characterized by a D614G point mutation that enhances transmissibility and has become the dominant strain in many countries since spring 2020.
In total, fourteen distinct lineages were identified amongst the 56 samples, representing multiple introductions of the virus to the Montana area.
Of the 56 samples, seven exhibited deletion to regions of the ORF7a encoding gene, one of which bore two additional point changes that were the result of community-acquired mutations within the same lineage.
A world-wide search of the available databases revealed 845 unique ORF7a gene variants, representing 189 unique ORF7a amino acid sequences that differ from the original Wuhan strain. These variants have appeared on every continent over the course of the pandemic and were not from a single lineage.
In comparing the translated ORF7a sequences, it was found that two-thirds bore a truncated C-terminus while the N-terminal half of the protein was preserved, with a third of these mutations occurring independently in two or more patients. Truncated ORF7a SARS-CoV-2 can, however, still be transferred between hosts, as some of the forms were confirmed to be from the same lineage.
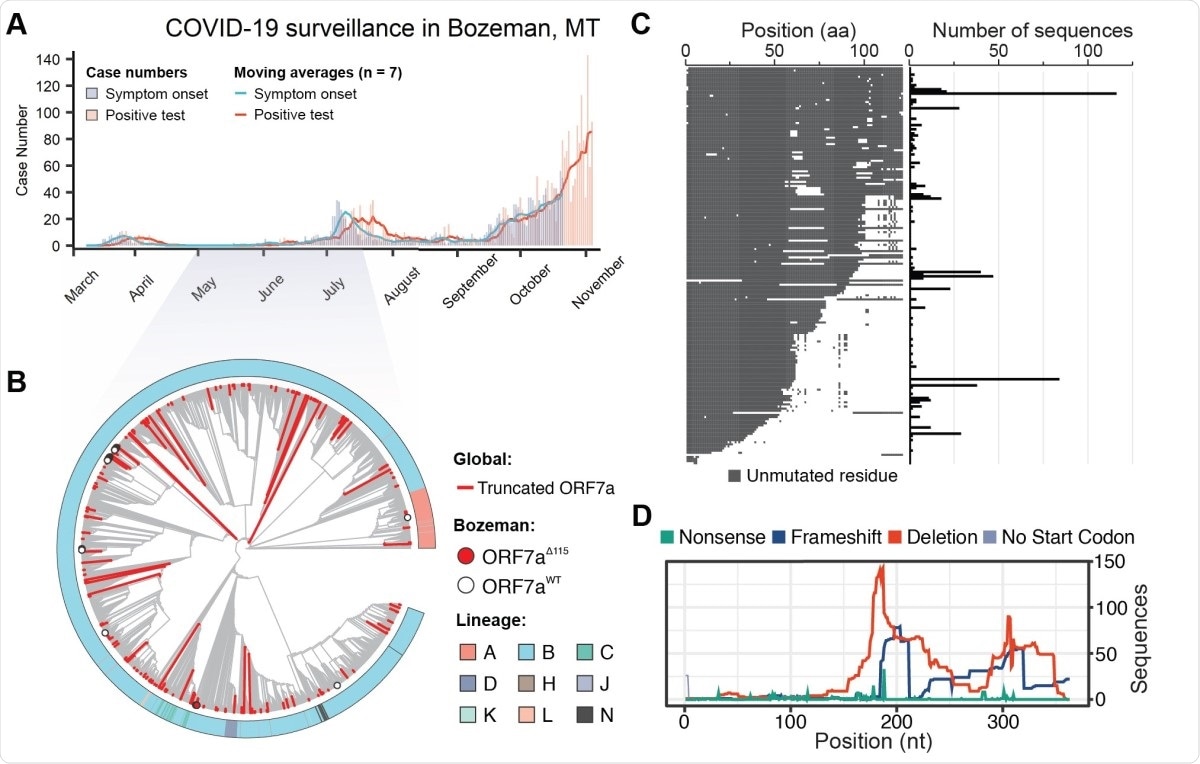
SARS-CoV-2 genomic surveillance identifies global reoccurrence of ORF7a truncations. A) Symptom onset (purple) and PCR-based SARS-CoV-2 test results (coral) for patients in Bozeman, Montana are shown with vertical bars. Seven day moving averages, shown with lines, were used to indicate epidemiological trends. B) Phylogenetic analysis of SARS-CoV-2 genomes sampled in Bozeman and globally. The tree was constructed from an alignment of 56 Bozeman samples and 4,871 genomes subsampled from GISAID. Subsampling was performed using Augur utility (Nextstrain) by selecting 10 genomes per country per month since the start of the pandemic. Red branches identify truncated ORF7a variants (n= 205) detected in the global data and merged into the alignment. The red dot highlights seven of the 56 ORF7a variants that were isolated in Bozeman between April and July (2020). White dots highlight 49 viral genomes isolated in Bozeman that have wild-type ORF7a sequences. C) Alignment of unique ORF7a protein sequences with five or more mutations as compared to the SARS-CoV-2 reference strain (Wuhan-Hu-1). Conserved residues are depicted in gray, while mutations and deletions are shown in white. The frequency of each genotype is shown on the right. D) Distribution of different mutations that occur along the ORF7a coding sequence.
What is the significance?
ORF7a is a transmembrane protein with an N-terminal immunoglobulin-like ectodomain that consists of two β sheets held together by two disulfide bonds. The mutated ORF7a bears an additional stop codon that eliminates these bonds, destabilizing the protein structure of ORF7a and impacting protein function.
This was evidenced further by fluorescent tagging of wild-type or mutated ORF7a in HEK293 cells, with the mutated form being delocalized from the cell membrane due to the destabilization. Viral replication rate was also assessed in the same cells by measurement of viral RNA using PCR, with wild type displaying a 960-fold increase over five days while the mutated virus showed only a 270-fold increase in population. This was confirmed to be an artifact of both lessened viral replication and viral egress.
The authors suggest, however, that the lower observed replication rate is a direct result of weakened suppression of host anti-viral immune response by ORF7a, as the protein is not essential to virus replication.
To observe this mechanism in further detail, the expression of 88 genes involved in interferon response was examined in HEK293 cells following exposure to wild-type or ORF7a-mutated SASR-CoV-2. Forty-two of these genes were noted to double in expression following exposure to wild type, while the mutated form doubled the expression of 55 distinct genes. Further investigation revealed that most of the former are involved in the downregulation of interferon processes, while the latter are more heavily involved in upregulation and strengthening of the host immune response.
SARS-CoV-2 genomic surveillance identifies naturally occurring truncations of ORF7a that limit immune suppression

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Nemudryi, Artem A. et al. (2021). SARS-CoV-2 genomic surveillance identifies naturally occurring truncations of ORF7a that limit immune suppression. medRxiv preprint server. doi: https://doi.org/10.1101/2021.02.22.21252253, https://www.medrxiv.org/content/10.1101/2021.02.22.21252253v1
- Peer reviewed and published scientific report.
Nemudryi, Artem, Anna Nemudraia, Tanner Wiegand, Joseph Nichols, Deann T. Snyder, Jodi F. Hedges, Calvin Cicha, et al. 2021. “SARS-CoV-2 Genomic Surveillance Identifies Naturally Occurring Truncation of ORF7a That Limits Immune Suppression.” Cell Reports 35 (9): 109197. https://doi.org/10.1016/j.celrep.2021.109197. https://www.cell.com/cell-reports/fulltext/S2211-1247(21)00543-X.