Even as vaccine rollouts are commencing in many countries worldwide, to combat the ongoing pandemic of coronavirus disease 2019 (COVID-19) – caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – new emerging variants undermine the possibility of completely arresting viral spread. This is due to their demonstrated partial resistance to vaccine-elicited antibodies and/or increased transmissibility, as well as increased virulence, depending on the variant.
A new study by a team of researchers in the UK and US describes the use of a computational tool that generates all possible single amino acid substitutions in SARS-CoV-2 and predicts their effects.
The researchers also compared their findings with expected findings based on the observed variant frequency and earlier experiments to identify more common variants with clinically significant effects. They also detect variants that may impact antibody efficacy but not other aspects of viral biology.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
The team has released their findings as a preprint on the bioRxiv* server.
Mutational effects
Among the variants of concern at present are B.1.1.7, B.1.351 and P.1, also called the UK, South African and Brazil strains, respectively.
The most common mutations are substitutions of one nucleotide for another. These affect both viral RNA structure and function, as well as protein sequence. The latter is the focus of the current study.
Changes in protein sequence can alter the fundamental structure, stability and activity of the protein, thus deeply impacting its biological function. Some examples are the E484K and the N439K mutations, which affect antibody binding and affinity for the human cell receptor for the virus, the angiotensin-converting enzyme 2 (ACE2).
Computational prediction tools make use of protein sequence and structure to assess the potential effects of mutations. If a variant appears at the same position in different species, it is more likely to become a fixed mutation, and this is the basis of tools like SIFT4G and EVCouplings.
Conversely, FoldX and Rosetta base their predictions on protein structure, modeling the changes caused by the substitutions on the energetics of the protein, and on its binding if such models are reported.
The high cost and technical challenges prevent the widespread use of such predictive tools, however.
New web-based tool
The current study is based on the Mutfunc web service developed to provide an interface where predictions for all potential variants of H. sapiens, M. musculus and S.cerevisiae (human beings, the housefly, and baker’s yeast, respectively) genomes have already been computed.
Application to SARS-CoV-2 mutations
This was used to analyze SARS-CoV-2 proteins, with a rich combination of factors that impact the predicted effects. These factors include the conservation of sequences across variants, the structure of the protein, known protein-protein interactions, phosphorylation sites, and variant frequencies. The service is available on the website sars.mutfunc.com, allowing the dataset to be searched or downloaded. The structures of various potential variants can also be seen on this interface.
The current study examines the validity of the predictions made using this tool as to variants of concern, their impact on the emergence of new strains, and on antibody-mediated neutralization of the virus.
They compared the scores obtained by SIFT4G predictions for variants known to exist at high frequencies and found that scores were lower for uncommon variants, as expected. With FoldX, lower scores were found for more frequently occurring variants.
Deep mutational scanning (DMS) on SARS-CoV-2 spike variants showed that harmful mutations had lower viral fitness measurements.
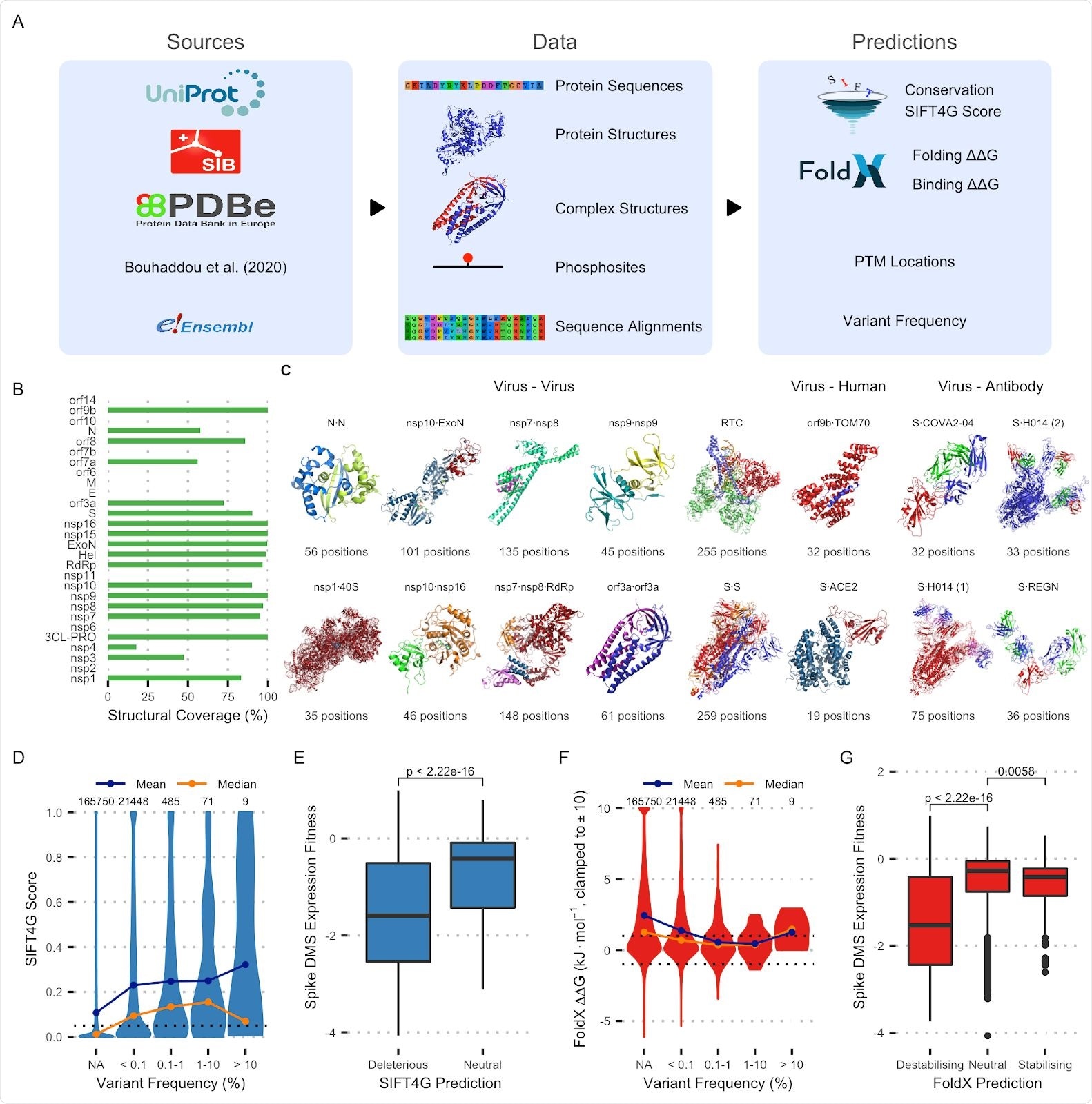
A: Data generation pipeline schematic. B: Percentage of residues covered by structural models for each protein. C: Protein complex structures currently included in the dataset. D: Distribution of SIFT4G scores for variants across frequencies. NA indicates variants that were not observed at all. The threshold for prediction being significant (0.05) is marked as is the number of variants in each category E: Distribution of Spike DMS variant expression fitness scores for variants predicted deleterious (< 0.05) or neutral (> 0.05) by SIFT4G. The p-value from a Wilcoxon signed-rank test is shown. F: Distribution of FoldX ΔΔG predictions for variants of varying frequencies. The thresholds for a variant being considered destabilising (1) and stabilising (-1) are marked. G: Distribution of Spike deep mutational scan variant expression fitness for variants predicted destabilising, neutral or stabilising by FoldX. P-values from Wilcoxon signed-rank tests are shown.
Protein-protein interfaces
Since protein-protein interactions are key to protein function, the researchers used FoldX scores on protein binding at these interfaces. They found that higher variant frequency is likely to be associated with less loss of stability.
Spike-ACE2 binding strength measured by DMS also showed lower scores, with variants expected to destabilize binding at this site. Thus, these predictions appear to offer valid insights on the impact of variants on viral biology.
The researchers also found that the nucleocapsid (N) dimerization interface, with its two beta-strands at the center, proved important for binding. These strands are liable to the greatest destabilizing impact following mutation.
The number of mutations in these regions is also higher compared to the rest of the interface. This makes this a very interesting region, with a combination of high-impact and frequent mutations.
Since there are multiple mutations in the same functional region and in different strains, they may confer a functional advantage.
High-frequency variants
The globally dominant D614G variant was predicted to prevent contact between this residue and the other spike subunit in the bound protein.
When used to predict the impacts of variants in the three currently circulating variant strains of concern mentioned above, the N501Y variant found in all three was predicted to adversely affect spike-ACE2 binding by destabilizing the interface. However, DMS shows increased binding instead.
This suggests [N501Y] changes the binding conformation in a way FoldX doesn’t accurately model and emphasizes that computational predictions are good at identifying variants that impact interfaces but do not always fully model the consequences.”
E484K, found in B.1.351 and P.1, and recently in some B.1.1.7 samples, was also predicted to have the same effect, along with others in both the UK and South African strains. Others, in the open reading frame (ORF) 8, in B.1.1.7 and B.1.351, may interfere with the immune response and vesicle transport.
ORF3 variants were predicted to induce apoptosis, with three of them potentially destabilizing the structure of the encoded dimeric ion channel, while one, the most frequent (Q57H), may hinder ion transport.
More than half of the nearly 50 variants that were found in a patient treated with convalescent plasma were associated with one or more significant predicted impact, correlating with their rise in frequency.
Immune evasion
Using all these methods, the researchers suggest that if a variant is expected to be neutral in its effects, based on binding at the ACE2-spike interface, sequence conservation and structure, but shows immune escape under experimental conditions, these are unlikely to be negatively selected and will therefore rise in frequency.
Such variants occur in two spike regions, both in the receptor-binding domain (RBD), the upper head, where most antibodies bind, and the base of the domain that connects it to the rest of the spike. Mutations at the latter are predicted to be universally deleterious, especially at positions 456 and 484, such as the E484K.
In fact, the latter has been predicted to reduce the stability of spike-H104 antibody binding, but both SIFT4G and FoldX predict it to be neutral. This indicates it is of deep concern. At other positions, the four variants with the greatest evasive potential are N501T, V503T, I472G and G485P, among which the first is predicted to be neutral in their effects. This makes them all the more concerning.
What are the implications?
Care must be taken when interpreting all results but particularly computational predictions,” say the researchers.
Firstly, these predictions come from mathematical models rather than being experimental results.
Secondly, the predictors only predict the deleterious nature of a change, with respect to protein function but not other viral features, including its infectivity or host immune response.
Thirdly, some features may be missing from the dataset, including some experimentally observed interactions with no structural basis, some phosphosites, and other post-translational modifications that are not even considered.
However, such studies can help search and identify the variants that may most probably affect protein function, both those that have been already identified and those that may emerge. The combination of predictions with experimental data on antibody escape mutations helped pick out those variants that were likely to preserve viral fitness but impact antibody binding.
The variants so identified are fit for deeper study and for continued monitoring. Meanwhile, other scientists may use the database on the server to complement experimental research and other analytical methods used to assess mutational impacts on virus-host interactions.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.