Each of the three approved vaccines against coronavirus disease 2019 (COVID-19) – Moderna, BioNTech-Pfizer and Oxford-Astrazeneca – generate forms of the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), allowing the immune system to respond with the appropriate antibodies. Since viruses are able to adapt and mutate at high rates, such vaccine cargoes may eventually become ineffective, and thus identifying highly conserved regions of the protein would be useful in the development of additional therapies.
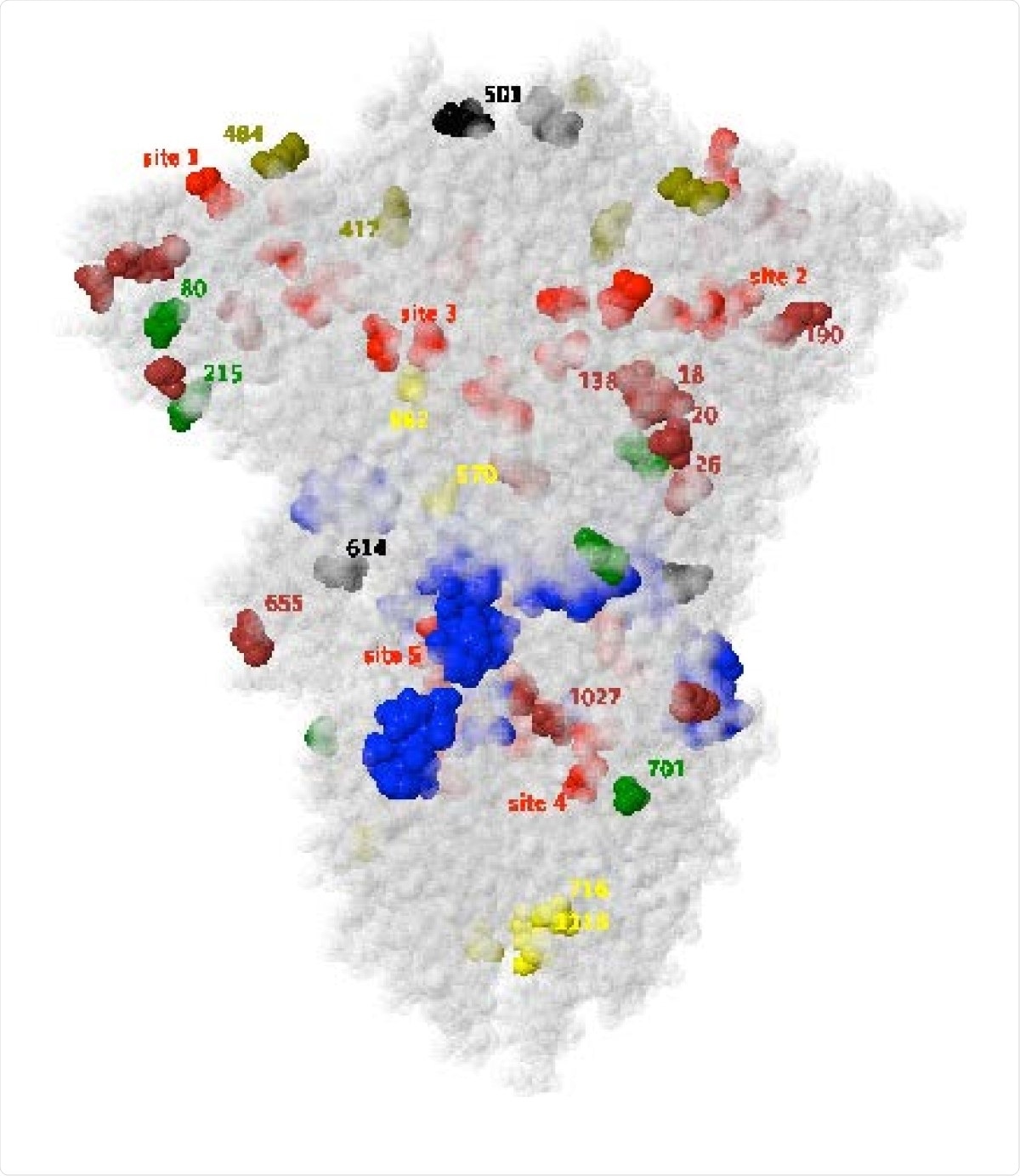
Mutated sites of the SARS CoV-2 spike glycoprotein for the variants of concern depicted in PDB file 7df3, which does not model residue 618. Yellow, brown and green residues are respective mutations for the U.K., Brazil and S. Africa variants, with olive residues 417 and 484 common to the latter two, and black residues 501 and 614 common to all three.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
A recent study by scientist Robert Penner at the Institut des Hautes Études Scientifiques in France utilizes free energy calculations to predict effective antiviral targets that are highly conserved, surface-accessible for interaction, and resistant to mutation. The study is available on the bioRxiv* server.
What is protein backbone free energy?
Backbone free energy (BFE) is a measure of the freedom of a section of a protein structure, mainly dependent on the number of hydrogen bonds between regions. Therefore, regions with high BFE are more likely to undergo a greater conformational change at some point, for example, while the virus spike protein is penetrating and fusing with the host cell.
Protein geometry is the main way in which immune cells recognize a pathogen, and independent proteins may take on a differing conformation than when part of a larger macromolecule. Therefore, vaccine cargoes must replicate the in-situ conformation of the living virus as closely as possible to elicit the ideal response.
High BFE is only evolutionarily conserved if essential to a particular function. In cases of conservation across several types of coronaviruses, the function must be critical to the proliferation and survival of the virus. Unfortunately, the high conformational mobility of these regions lessens their attractiveness as viral targets, as targeting the changing 3D structure is made more difficult. Low BFE sites, in contrast, are more static in conformation, and likely fold similarly in isolation. Additionally, as low BFE sites are surrounded by a large number of hydrogen bonds, any mutations in the BFE site should preserve antigen structure. Any mutations here will not be critical to function and thus be adopted wide-scale by evolutionary pressures.
How can low-BFE sites be exploited?
Highly conserved low BFE sites, therefore, are relatively rare. As they are conserved across multiple strains of coronavirus, they must play an essential function, though they are static and therefore make good, stable drug targets. The researcher hypothesizes that immobilization of passive low BFE sites may interfere with the activity of nearby high BFE sites that are critical to function.
Several SARS-CoV-2 variants of concern have been identified with mutations to the spike protein that allow for conformational changes beyond those observed in wild type. The greater conformational freedom has been implied to be involved in the greater transmissibility seen in these strains. The group obtained 29 structural files for SARS-CoV-2 spike proteins from various variants of concern. Mutant residues were identified, and the BFE was compared, with around half of the mutants exhibiting a higher BFE than wild-type counterparts due to fewer hydrogen bonding interactions.
The functional purpose of certain residues can be inferred by their conformational changes at specific pH. For example, one mutant residue was noted to have a high BFE at low pH, indicating the need for greater conformational activity in more acidic environments, such as when entering cells via the endocytotic pathway.
Four nine-base-pair long passive sites of interest that were accessible on the surface of the SARS-CoV-2 spike protein were identified that were common to each of the 29 variants of concern, each having a low BFE and each located upstream of a large active binding supersite. These sites could play a role in the critical functioning of the SARS-CoV-2 spike protein, and exploiting these sites may play a future role in the development of reliable and long-lasting COVID-19 vaccines.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources