New research suggests Syrian golden hamsters could be a potentially useful preclinical model for testing vaccine immunity against the acute respiratory syndrome coronavirus 2 (SARS-CoV-2) B.1.17 and B.1.351 variant.
An international team of scientists found evidence that the course of infection from both variants replicated in hamsters, along with a similar immune system profile observed in humans.
Their findings could help study the effectiveness of current vaccines undergoing modifications to fight against the new variants and future vaccines and treatments.
The researchers write:
"In an urgent need to characterize current VoC and in anticipation of future needs, the robust hamster model described here will allow to preclinically asses (i) virus transmission, (ii) vaccine efficacy, and (iii) evaluation of pharmacological interventions that target B.1.1.7 and B.1.351 as well as expected future VoC."
The study "Comparative infectivity and pathogenesis of emerging SARS-CoV-2 variants in Syrian hamsters" is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
Targeting most prevalent variants of concern
The B.1.1.7 variant was first discovered last fall in London and rapidly became the dominant strain in the United Kingdom (UK).
The B.1.351 variant was first reported in South Africa and contains mutations on the spike protein that allow it to be resistant against neutralizing antibodies created from either natural infection or vaccines. An observed decrease in vaccine immunity has been linked to the E484K mutation in the B.1.351 variant and more recent UK sub-lineages.
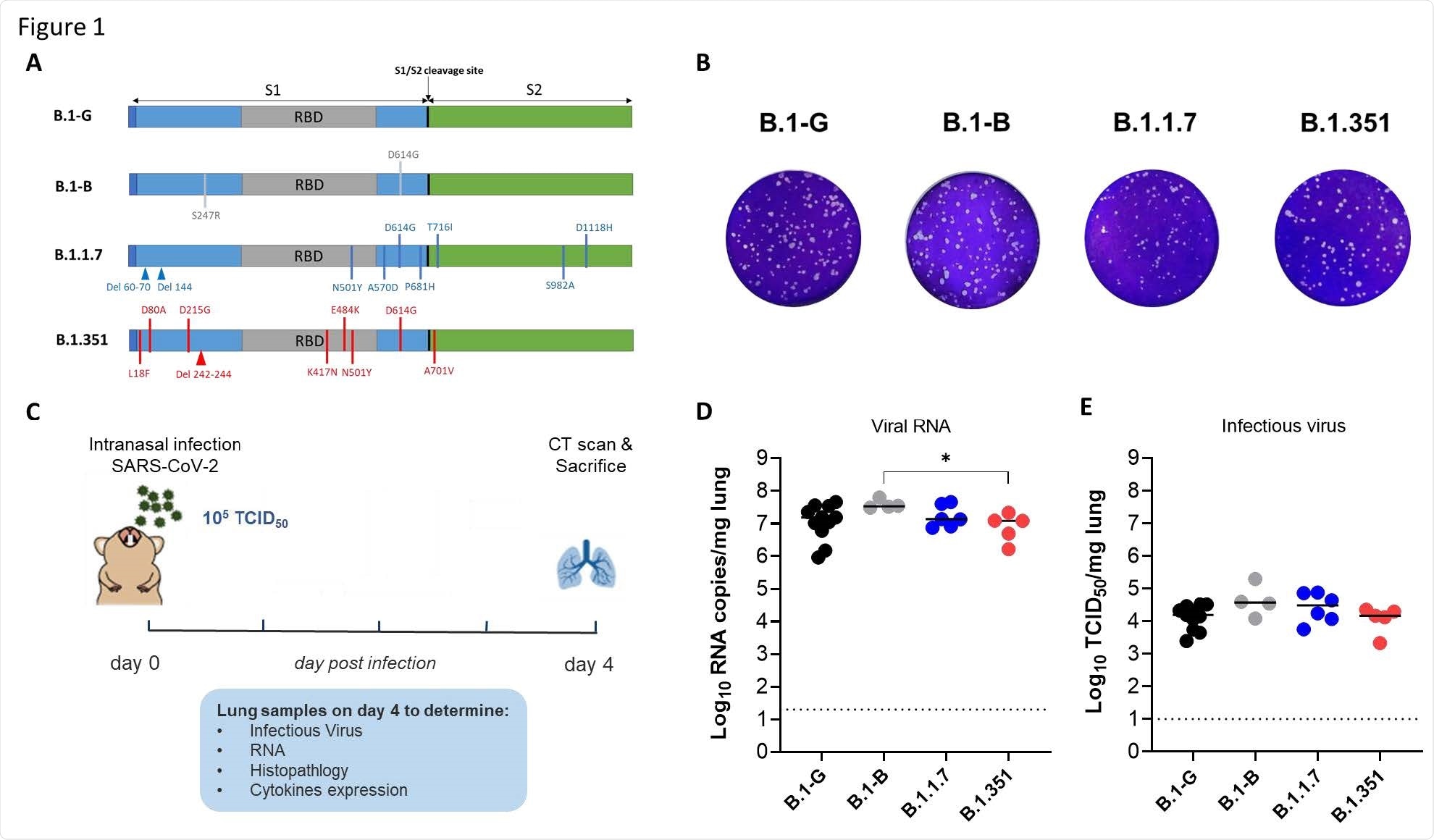
Characterization of the in vitro and in vivo replication of different SARS-CoV-2 variants. (A) Graphical representation for the SARS-CoV-2 spike gene showing the genotypic difference between the B.1-G, B.1-B, B.1.1.7 and B.1.351 SAR-CoV-2 variants. (B) Plaque phenotype of B.1-G, B.1-B, B.1.1.7 and B.1.351 SAR-CoV-2 variants in Vero E6 cells. (C) Set-up of the Syrian hamsters infection study. (D) Viral RNA levels in the lungs of hamsters infected with 105 TCID50 of B.1-G (n=11), B.1-B (n=4), B.1.1.7 (n=6) or B.1.351 (n=5) SAR-CoV-2 variants on day 4 post-infection, p.i are expressed as log10 SARS-CoV-2 RNA copies per mg lung tissue. Individual data and median values are presented. (E) Infectious viral loads in the lungs of hamsters infected with the different SARS-CoV-2 variants at day 4 pi are expressed as log10 TCID50 per mg lung tissue. Individual data and median values are presented. Data were analyzed with the Mann−Whitney U test. *P < 0.05.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Animal model setup
The researchers intranasally infected 6-8-week-old female Syrian hamsters with 50 microliters of four strains. This included basal lineages B.1-G or B.1-B, the B.1.17 variant, or the B.1351 variant. There were approximately 4-8 hamsters per group.
Four days after infection, the team looked at the hamsters' lung tissue to measure post-infection status, SARS-CoV-2 replication, pathology, and inflammation levels.
Results
All four strains showed viral RNA load and infectious virus titers in the lung tissue. However, when the B.1.1.7 and B.1.351 variant was given at a lower dose, they showed similar viral RNA loads and infectious virus titers. The researchers suggest the hamster results align with the high susceptibility for infection observed in both variants.
Looking at variants' effects on lung pathology, the researchers observed similarities in pulmonary infiltrates and bronchial dilation in the hamsters infected with the B.1.1.7 and B.1.351 variant.
There was also evidence of peri-bronchial inflammation and bronchopneumonia in surrounding alveoli, similar to the bronchopneumonia reported in patients with COVID-19 infection.
The team also found that hamsters' lung tissue showed a similar cytokine profile to the one observed with COVID-19 infection in humans.
"Infection with either of the 203 four strains resulted in an up-regulation of IL-6, IL-10, IFN-λ, IFN-γ, IP-10, MX-2, and TNF-α 204 expression in the range of 10- to 1000-fold compared to non-infected hamsters," write the researchers.
The B.1.1.7 variant specifically showed the most upregulation for IL-6, IL-10, and IFN-γ — but not MX-2 expressions. The cytokine expression levels did not change when the viral dosage was reduced.
However, ACE2 receptor expression remained virtually unchanged in all four strains. A small upregulation of ACE2 receptor expression was only observed in hamster lungs infected with basal B.1-G and B.1-B lineage. For this reason, the researchers say the hamster model currently does not explain the epidemiology, transmission, infectivity, or virulence of people infected with the SARS-CoV-2 variants.
Researchers suggest the small number of animals per group may have contributed to the lack of differences.
Evolution of COVID variants
The driving force behind the emergence of new SARS-CoV-2 strains remains unknown.
"The emergence of future VoC may be driven by any of the following factors: by random selection (founder effects), fitness at the population level (favoring transmission), or viral escape under host immune pressure (antigenic drift), or development of drug resistance under future antiviral therapy. Whatever the cause, as a consequence, any upcoming VoC may spark future COVID-19 epidemics."
However, the researchers note that genomic sequencing of viral RNA from the infected lungs of hamsters did not show further sequence evolution in the spike gene 4 days post-infection.
Additionally, no convergent evolution for higher virulence or significant change in phenotype was observed in both variants.
Based on the findings, the researchers suggest hamsters could be a durable preclinical model to replicate the human experience when infected with either the B.1.1.7 and B.1.351 variants.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Abdelnabi R, et al. Comparative infectivity and pathogenesis of emerging SARS-CoV-2 variants in Syrian hamsters, bioRxiv, 2021. doi: https://doi.org/10.1101/2021.02.26.433062, https://www.biorxiv.org/content/10.1101/2021.02.26.433062v1
- Peer reviewed and published scientific report.
Abdelnabi, Rana, Robbert Boudewijns, Caroline S. Foo, Laura Seldeslachts, Lorena Sanchez-Felipe, Xin Zhang, Leen Delang, et al. 2021. “Comparing Infectivity and Virulence of Emerging SARS-CoV-2 Variants in Syrian Hamsters.” EBioMedicine 68 (June): 103403. https://doi.org/10.1016/j.ebiom.2021.103403. https://www.thelancet.com/journals/ebiom/article/PIIS2352-3964(21)00196-1/fulltext.