Researchers from California in the United States have conducted a study showing that important adaptive immune responses to infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are not affected by four recently emerged variants of the virus.
The SARS-CoV-2 virus is the agent responsible for the coronavirus disease 2019 (COVID-19) pandemic that continues to pose a threat to global public health and has now claimed the lives of more than 2.55 million people.
Several studies have demonstrated that mutations present in recently emerged variants of SARS-CoV-2 confer resistance to neutralizing antibodies (the humoral immune response) in sera from convalescent and vaccinated individuals.
This has highlighted the need to better understand adaptive immune responses to the virus, say researchers from La Jolla Institute for Immunology, the Craig Venter Institute in La Jolla, and the University of California in San Diego.
“It is important to address whether CD4+ and CD8+ T cell responses are also affected because of the role they play in disease resolution and modulation of COVID-19 disease severity,” writes Alessandro Sette and colleagues.
Now, the team has conducted a comprehensive analysis showing that CD4+ and CD8+ T cell responses in convalescent individuals and recipients of the Pfizer-BioNTech and Moderna COVID-19 vaccines are not substantially affected by mutations found in four recently emerged SARS-CoV-2 variants.
The variants tested included the B.1.1.7, B.1.351, P.1 and CAL.20C variants that have emerged over recent months in the UK, South Africa, Brazil and California, respectively.
“The data provide some positive news in light of justified concern over the impact of SARS-CoV-2 variants of concern on efforts to control and eliminate the present pandemic,” says the team.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
More about the variants of concern
The recent emergence of several SARS-CoV-2 variants containing multiple mutations has important implications for the future control of the COVID-19 pandemic.
The mutations causing the most concern are those found in the viral spike protein. This spike is the main structure SARS-CoV-2 uses to bind to and infect host cells and is also the protein on which several COVID-19 vaccines are based, including the Pfizer-BioNTech BNT162b2 vaccine and the Moderna mRNA-1273 vaccine.
Several of these mutations have been shown to directly affect host cell receptor binding affinity, which may increase infectivity, viral load and transmissibility.
Furthermore, some mutations identified in spike regions that are bound by neutralizing antibodies have been shown to confer resistance to neutralization by sera from convalescent and vaccinated individuals.
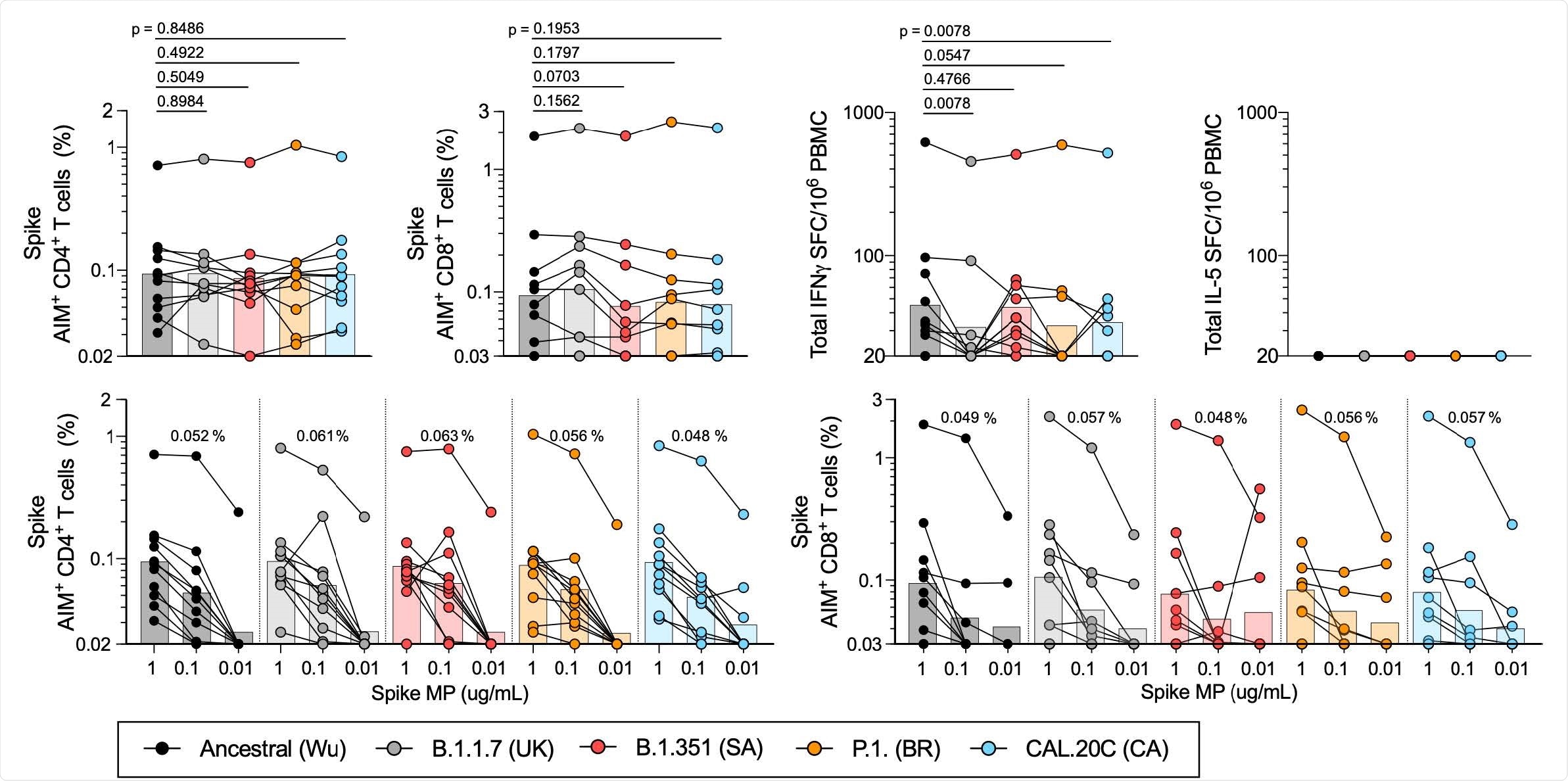
T cell responses of COVID-19 convalescent individuals against SARS-CoV-2 Spike for the different variants. PBMCs of COVID-19 convalescent individuals (n=11) were stimulated with the Spike MPs corresponding to the ancestral reference strain (Wu, black) and the B.1.1.7 (UK, grey), B.1.351 (SA, red), P.1 (BR, orange) and CAL.20C (CA, light blue) SARS-CoV-2 variants. A) Percentages of AIM+ (OX40+CD137+) CD4+T cells. B) Percentages of AIM+ (CD69+CD137+) CD8+ T cells. C) IFNγ spot forming cells (SFC) per million PBMCs D) IL-5 SFC per million PBMCs. Paired comparisons of Wuhan S MP versus each of the variants were performed by Wilcoxon test and are indicated by the p values in panels A-C. The data shown in panels A and B are plotted to show the Spike MPs titration (1 μg/mL, 0.1 μg/mL, 0.01 μg/mL) for CD4+ (E) and CD8+ (F) T cells in each SARS-CoV-2 variant and the geometric mean of the 0.1ug/mL condition is listed above each titration. In all panels, the bars represent the geometric mean.
What about T cell responses?
A growing body of evidence suggests that CD4+ and CD8+ T cell responses – essential elements of adaptive immunity – modulate COVID-19 disease severity in humans and reduce viral load in non-human primates.
Multiple studies have shown that SARS-CoV-2 infection induces robust CD4+ and CD8+ T cell memory and several COVID-19 vaccines have been shown to elicit CD4+ and CD8+ T cell responses.
“It is therefore key to address the potential impact of SARS-CoV-2 variants mutations on T cell reactivity,” says Sette and colleagues.
What did the researchers do?
The researchers tested how the variants of concern B.1.1.7 (UK), B.1.351 (South Africa), P.1 (Brazil), and CAL.20C (California) impact on SARS-CoV-2-specific CD4+ and CD8+ T cell responses using blood samples taken from COVID-19 convalescent individuals previously infected with the ancestral viral strain (detected in Wuhan China).
They also tested samples collected from recipients of the Pfizer-BioNTech or Moderna vaccine approximately 14 days following administration of the second dose.
The team used activation-induced marker assays to measure CD4+ and CD8+ T cell responses to peptide pools spanning the Spike antigen of the ancestral strain and the four variant strains.
What did they find?
The convalescent samples exhibited good CD4+ and CD8+ T cell reactivity to pools of peptides spanning the spike of the ancestral Wuhan strain and the four variant strains.
No significant differences were observed in CD4+ and CD8+ T cell reactivity to the pool of ancestral spike peptides and the four variant pools.
“These experiments suggest that memory CD4+ or CD8+ T cells from individuals that have been infected with the ancestral SARS-CoV-2 strain recognize the ancestral reference strain and the variant genome-wide sequences with similar efficiency,” writes Sette and the team.
Similarly, no significant differences were observed in CD4+ and CD8+ T cell reactivity to ancestral versus variant spike peptides among vaccinated individuals, with the exception of the B.1.351 peptide pools.
Vaccinated samples tested against the B.1.351 spike peptides showed mild decreases of 29% in CD4+ T cell reactivity and 33% in CD8+ T cell reactivity.
What did the authors conclude?
The researchers say they found negligible effects on both CD4+ or CD8+ T cell responses to all four variants investigated.
“Overall, the results demonstrate that CD4+ and CD8+ T cell responses in convalescent COVID-19 subjects or COVID-19 mRNA vaccinees are not substantially affected by mutations found in the SARS-CoV-2 variants,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Sette A, et al. Negligible impact of SARS-CoV-2 variants on CD4+ and CD8+ T cell reactivity in COVID-19 exposed donors and vaccinees. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.02.27.433180, https://www.biorxiv.org/content/10.1101/2021.02.27.433180v1
- Peer reviewed and published scientific report.
Tarke, Alison, John Sidney, Nils Methot, Esther Dawen Yu, Yun Zhang, Jennifer M. Dan, Benjamin Goodwin, et al. 2021. “Impact of SARS-CoV-2 Variants on the Total CD4+ and CD8+ T Cell Reactivity in Infected or Vaccinated Individuals.” Cell Reports Medicine 2 (7). https://doi.org/10.1016/j.xcrm.2021.100355. https://www.cell.com/cell-reports-medicine/fulltext/S2666-3791(21)00204-4.