The ceaseless pandemic of coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 still has a major impact on human health globally, as it can give rise to severe disease and accompanying long-term health consequences.
Recently, there was a surge of unusual public health events that may result from variants of SARS-CoV-2. For example, the World Health Organization (WHO) routinely monitors whether these viral variants result in changes in transmission patterns, disease severity, or vaccine efficacy.
One notable SARS-CoV-2 variant is the infamous B.1.1.7 strain, which was initially detected in the United Kingdom in September 2020 and has since spread to more than fifty countries. In South Africa, another variant emerged independently of B.1.1.7, and there is also a Brazil variant known as P.1.
The problem is that mutations in the aforementioned variants may reduce recognition by antibodies elicited by natural infection with parental or original strains. This is why a large group of researchers, led by Dr. George Kassiotis from the Francis Crick Institute and the Imperial College London, decided to examine the degree of immunogenicity and heterotypic immunity the new variants may afford.
Appraising inhibitory concentrations of human sera
The sera for this study has been collected from 29 patients that were admitted to University London College Hospital for unrelated reasons, but with subsequently confirmed B.1.1.7 infection. The majority (i.e., 79%) of these patients displayed relatively mild COVID‐19 symptoms, while a smaller fraction of 21% remained asymptomatic.
To appraise potentially reduced antibody recognition and its functional consequence, the researchers have measured the half-maximal inhibitory concentration (usually abbreviated as IC50) of D614G and B.1.1.7 sera, utilizing an in vitro neutralization process of authentic Wuhan, B.1.1.7 (UK variant) and/or B.1.351 (South African variant) SARS-CoV-2 isolates.
Overall, IgG, IgM and IgA antibodies to the spike glycoprotein of the Wuhan strain – but also of variants D614G (this mutated version basically replaced the initial SARS-CoV-2 strain found in China), B.1.1.7 or B.1.351 – were expressed on human HEK293T cell line and subsequently detected by a flow cytometry‐based technique.
Different cross-neutralization patterns
This study has shown that the infection with D614G induced substantial and long-lasting cross-neutralization of the B.1.1.7 variant; however, the reverse was not true. In other words, the neutralization of the parental Wuhan strain by B.1.1.7 sera was significantly reduced in comparison to the neutralization properties of the infecting B.1.1.7 variant
Similar findings were independently found in a recent study with a rather small number of B.1.1.7 convalescent sera. Such a unidirectional pattern of cross‐reactivity implies that the likely culprit for the emergence of B.1.1.7 was not the process known as 'antibody escape.'
Furthermore, the difference in the cross-neutralization drop was also statistically significant. More specifically, both D614G and B.1.1.7 sera demonstrated significantly lower neutralization of the B.1.351 variant with a fold change of -8.2 and -7.7, respectively.
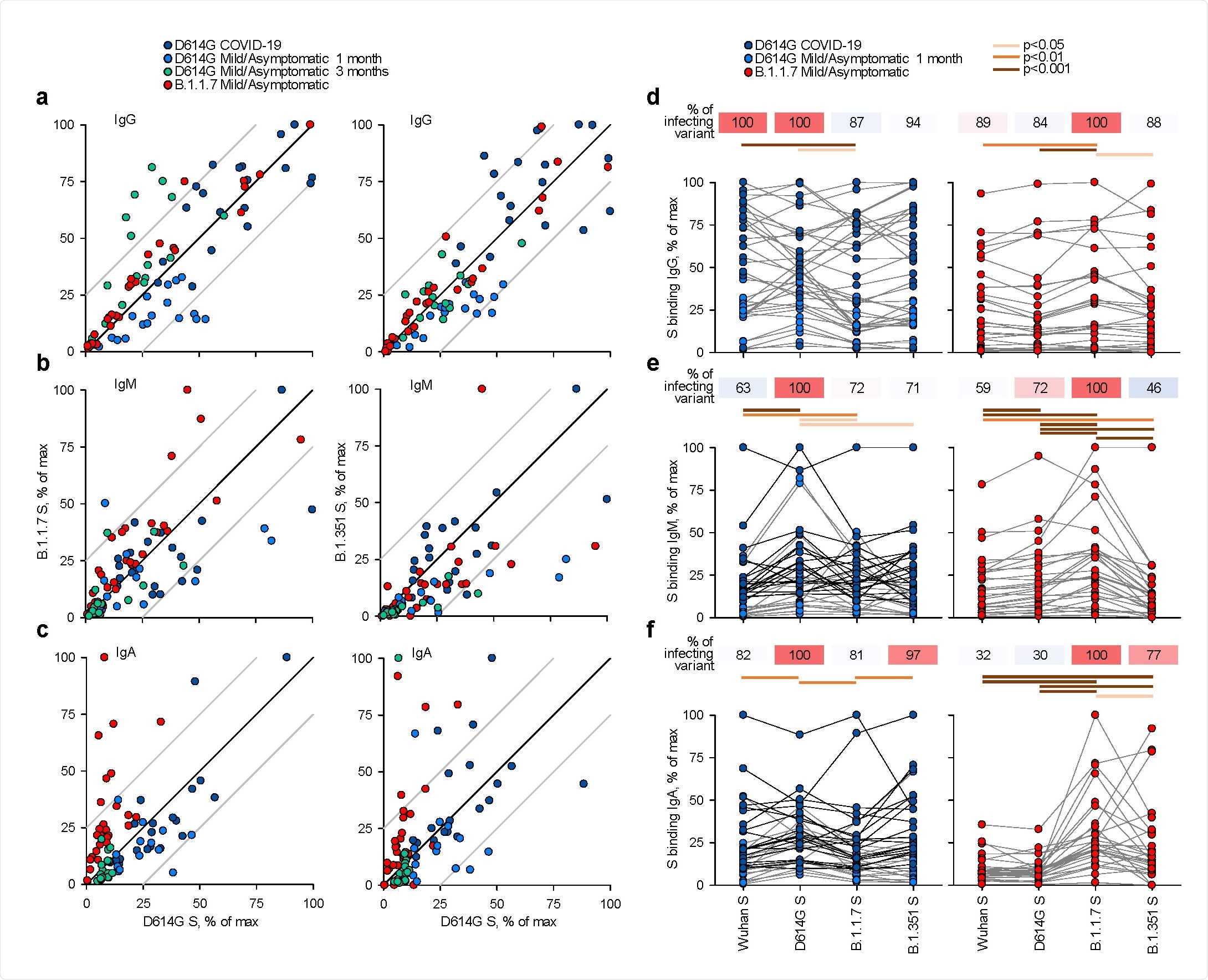
Recognition of distinct SARS‐CoV‐2 spike glycoproteins by antibodies in D614G and B.1.1.7 sera. a‐c, Correlation of IgG (a), IgM (b) and IgA (c) antibody levels to D614G and B.1.1.7 or B.1.351 spikes in the indicated groups of donors infected either with the D614G or B.1.1.7 strains. Each symbol represents an individual sample and levels are expressed as a percentage of the positive control. Black lines denote complete correlation and grey lines a 25% change in either direction. d‐f, Comparison of IgG (d), IgM (e) and IgA (f) antibody levels to the indicated spikes in groups of donors acutely infected either with the D614G or B.1.1.7 strains. Connected symbols represent individual donors. Numbers above the plots denote the average binding to each spike, expressed as a percentage of binding to the infecting spike.
The era of multivalent vaccines?
In a nutshell, these results suggest that natural infection with each strain of SARS-CoV-2 prompts the formation of antibodies that recognize the infecting strain most robustly, with various degrees of cross-recognition of other strains.
"Although a quantifiable correlation between in vitro neutralization of infectious SARS‐CoV‐2 and in vivo protection from SARS-CoV-2 infection or severe COVID-19 remains to be defined, the reduced neutralization of other SARS‐CoV‐2 strains by B.1.1.7 sera would suggest that the recent wave of global B.1.1.7 infections may not completely protect against re-infection with other SARS‐CoV‐2 strains", say study authors in this bioRxiv paper.
The degree of heterotypic immunity will have to be considered when selecting spike variants as vaccine candidates, with B.1.1.7 spike demonstrating lower potential when directly compared to other variants. In conclusion, such antigenic variation linked to SARS‐CoV‐2 evolution means that we may turn to the use of multivalent vaccines.
Source:
- Centers for Disease Control and Prevention (CDC). (2021). Science Brief: Emerging SARS-CoV-2 Variants. https://www.cdc.gov/
Journal reference: