While there has been a lot of research aimed at determining who is most susceptible to developing severe coronavirus disease 2019 (COVID-19) infection, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), there is a limited understanding of the biological mechanisms driving severe disease.
New research by a team of scientists at University Hospital RWTH Aachen in Germany and Erasmus Medical Centre in The Netherlands characterized changes in CD8+ T cells in response to SARS-CoV-2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The authors write:
In summary, this data suggests that CD8+ T cell response to SARS-CoV-2-derived epitopes preferentially leads to short-lived, terminal effector differentiation, and a hyperactivated phenotype in severe COVID-19, while epitope-binding CD8+ effector T cells in mild COVID-19 preserve the ability to develop into memory cells after clearance of the viral infection.”
Understanding the pathogenesis of severe COVID-19 could help adjust vaccine strategies and develop additional targeted therapies for patients.
The study “Dissecting CD8+ T cell pathology of severe SARS-CoV-2 infection by single-cell epitope mapping” is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
Infection severity elicits CD8+ T cell response
Researchers profiled the CD8+ T cell response during mild and severe SARS-CoV-2 infection. They combined scRNA and TCR-sequencing with CITE-seq antibodies and peptide-MHC class I Dextramer reagents to perform the study.
The researchers found CD8+ effector T cell differences in expressing an effector phenotype in people with mild and severe infection. Compared to mild infections, patients with severe infection showed characteristics of higher activation, indicating hyperactivation. They also observed more reduced characteristics of functional differentiation of the effector phenotype in severe than in mild infections.
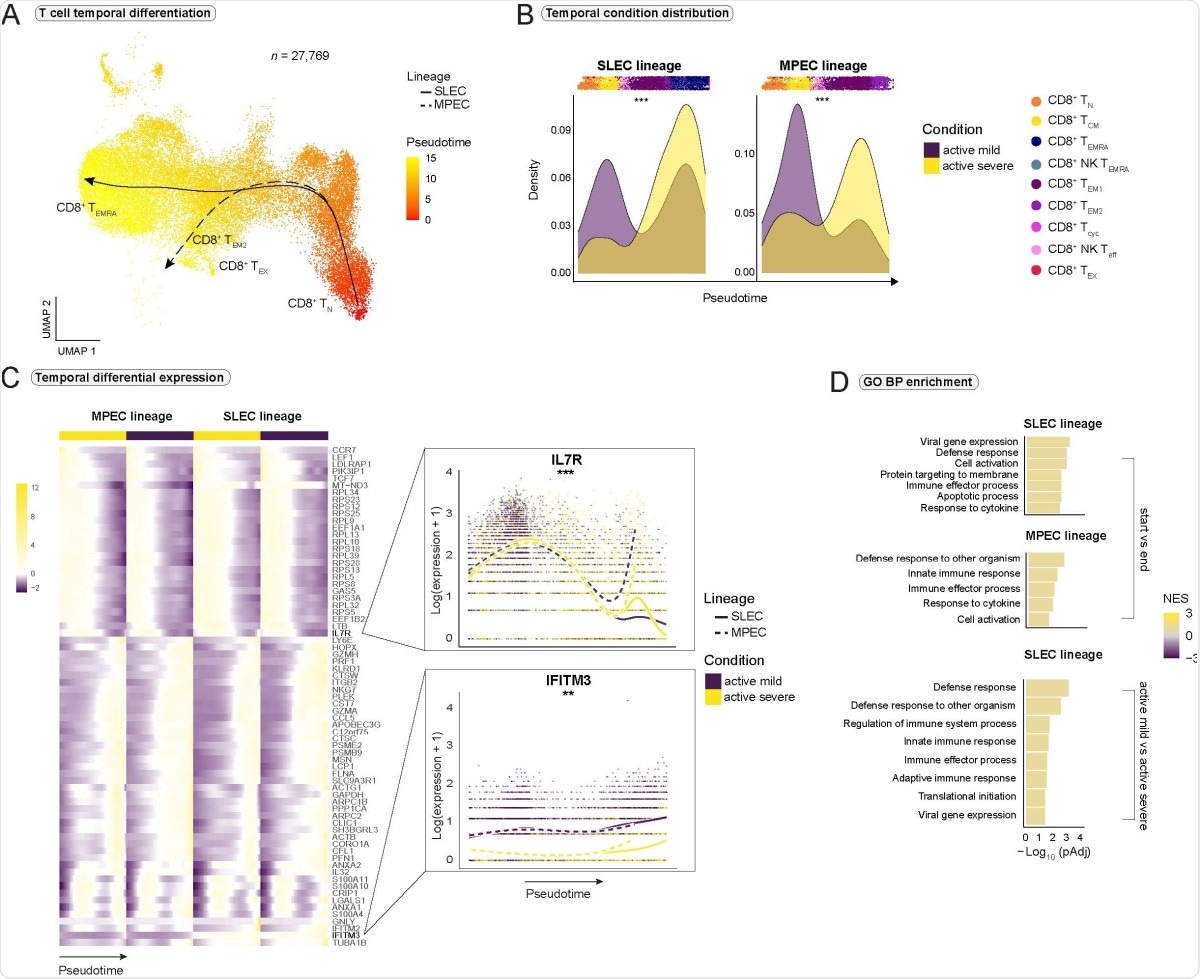
Impaired CD8+ T cell differentiation and exhaustion in severe COVID-19. (A) Pseudotimes and estimated trajectories projected onto the integrated UMAP of cell types likely to have their origin in naive CD8+ TN cells. (B) Temporal distribution of cell density for the active mild and severe conditions across pseudotime. Shifts in distribution between the conditions for the short-lived effector cells (SLEC) and memory precursor effector cells (MPEC) lineage were tested with the Kolmogorov-Smirnov method (MPEC: D = 0.310, p < 2e-16, SLEC: D = 0.402, p < 2e-16). Supplementary Fig. 4A is referred to for distribution of healthy and recovered conditions. (C) Heatmap depicts differentially expressed genes between the progenitor and differentiated cell populations across pseudotime (start vs end). Smoothed expression of two selected genes is shown with the y-axis on natural logarithmic scale. An extended panel of genes is reported in Supplementary Fig. 4. (D) Significantly enriched biological process (BP) gene sets from the Gene Ontology (GO) database. Gene set enrichment analysis was based on the genes from the start vs end test (upper panel) and condition tests between active mild and severe COVID-19 (lower panel) with tradeSeq. Normalized enrichment score = NES. *** = p-value < 0.001, ** = p value < 0.01.
When the team investigated clonal expansion of CD8+ T cells, they found a higher proportion of hyperexpanded clones in severe disease than mild disease.
A relevant CD8+ T cell epitope that could drive clonal expansion was identified as S protein-derived peptide A*0101 WTAGAAAYY.
Compared to severe COVID-19, WTAGAAAYY epitope-binding cells decreased gene expression for transcription factors and cytokine receptors associated with T cell memory formation. A cell-cell interaction analysis also showed people with severe disease had ligand-receptor impairments in their CD8+ T cells, which suggests a weakening of long-term T-cell formation.
These results might indicate a lower potential for formation of a long-lived SARS-CoV-2 specific CD8+ T cell memory in severe COVID-19,” concluded the team.
In contrast, researchers found increased gene expression associated with activation and exhaustion in epitope-binding cells in severe COVID-19 infection.
The results correspond with previous studies that observed a population of exhausted CD8+ T cells in patients who recovered from severe COVID-19. The findings suggest an inflammatory environment could favor the development of this type of exhausted T cells.
Indeed, the results did show CD8+ effector T cells having difficulties responding to interferon stimulations. Severe infections showed more downregulation of IFN-stimulated genes than mild infections.
The researchers attribute the T cell’s dysregulated response to IFN stimulation and to an impaired JAK-STAT signaling pathway.
Across all impairments in response to IFN-stimulated genes, IFITM3-expression was significantly reduced in all CD8+ T cell subtypes in severe COVID-19 infection.
However, in mild COVID-19, JAKSTAT-signaling seems to be sufficient to mount significantly higher expression levels of IFN-stimulated genes compared to severe disease. Interestingly, these findings were reversed when comparing WTAGAAAYY epitope-binding cells between severe and mild COVID-19, indicating that epitope-recognition and TCR stimulation are responsible for these changes.”
Non-activated CD8+ T cells involve activation of STAT1/2 signaling to upregulate IFN-stimulated genes. However, the researchers found an increase in STAT1 and decreased STAT4 expression in the epitope-binding CD8+ T cell population in severe disease.
A possible explanation for exhaustion in the CD8+ T cell population is the dysregulation of NFAT and AP-1, which rely on karyopherins for nuclear translocation. Given the T cells show exhaustion and an increase in cytotoxic effector molecules simultaneously, the researchers suggest this could be a potential mechanism of action for the exhaustion and subsequent hyperactivation.
Future work needed
The researchers note more research is needed to understand if the virus entering CD8+ T cells is the reason behind dysregulated responses for interferon stimulation and JAK-STAT signaling.
It also remains unknown whether reduced expression of IFITM3 contributes to viral infection in CD8+ T cells and, subsequently, increasing severity of COVID-19 illness.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Schreibing F, et al. Dissecting CD8+ T cell pathology of severe SARS-CoV-2 infection by single-cell epitope mapping. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.03.03.432690,https://www.biorxiv.org/content/10.1101/2021.03.03.432690v1.article-info
- Peer reviewed and published scientific report.
Schreibing, Felix, Monica T. Hannani, Hyojin Kim, James S. Nagai, Fabio Ticconi, Eleanor Fewings, Tore Bleckwehl, et al. 2022. “Dissecting CD8+ T Cell Pathology of Severe SARS-CoV-2 Infection by Single-Cell Immunoprofiling.” Frontiers in Immunology 13 (December). https://doi.org/10.3389/fimmu.2022.1066176. https://www.frontiersin.org/articles/10.3389/fimmu.2022.1066176/full.