Antibodies are central to infection control and prolonged immunity. Key tools in biomedical research, antibodies are also used to diagnose human diseases and detect adaptive immunity to past infections with particular pathogens.
While existing methods to discover and generate antibodies are successful, they also have important limitations, such as hard targets (short-lived, homologous, or unstable) and time-consuming and cost-intensive processes, often yielding high-affinity antibodies but with poor biophysical properties.
To overcome these challenges, computational studies coupled with biophysics and biotechnology have attracted growing attention. Particularly, the de novo computational designing of antibody binding has been most successful in synergy with in vitro affinity maturation, and in particular when applied to mini-proteins. These methods reduce the time and cost of antibody discovery while enabling the facile and precise targeting of specific epitopes.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
In a recent bioRxiv* preprint, a new study described a novel fragment-based method to design antibody complementarity determining region (CDR) loops.
In this study, the interdisciplinary team demonstrated the ability to design de novo antibodies that can bind to pre-selected epitopes of interest; they designed these on standard laptops, using a computer code.
The designed CDRs were then grafted onto antibody scaffolds and further optimized computationally for solubility and conformational stability. They found the binding KD values down to the nanomolar range, without any in vitro affinity maturation.
The researchers designed and tested six single-domain antibodies that targeted different epitopes on three antigens, including the receptor-binding domain (RBD) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein. SARS-CoV-2 being the pathogen responsible for the coronavirus disease 2019 (COVID-19) pandemic.
Interestingly, the researchers showed that a high-resolution input of the antigen structure is not required. The method here also yields similar predictions even when the input is a crystal structure or a computer-generated model. Therefore, this enables a rapid generation of lead antibodies binding to pre-selected epitopes.
De novo CDR-design strategy
The researchers compiled from the non-redundant Protein Data Bank (PDB) a database of CDR-like fragments and corresponding antigen-like regions.
The structure of the input epitope was first fragmented, followed by each epitope fragment compared to the antigen-like regions to identify the ones with compatible backbone structure and a similar sequence. The resulting CDR-like structures were then rotated to match the epitope's orientation by superimposing each antigen-like region.
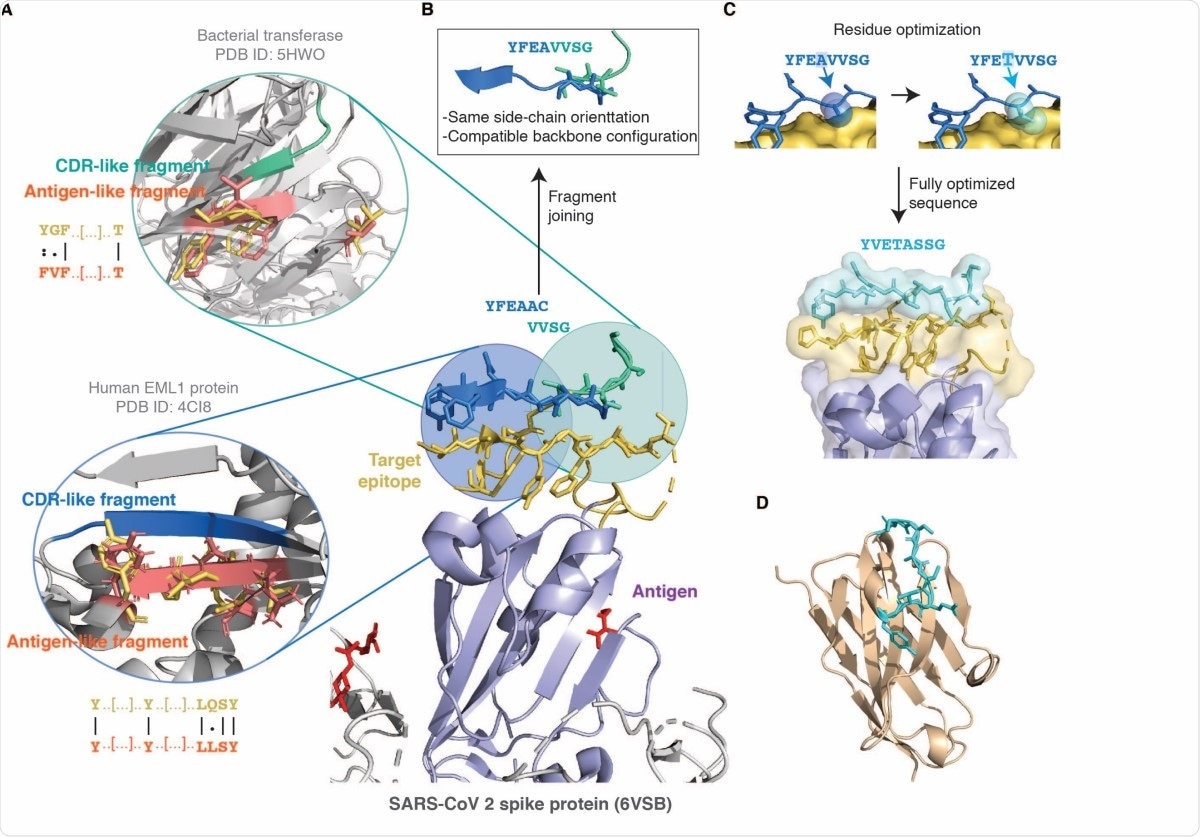
Schematic illustration of the combinatorial structure-based CDR design strategy introduced in this work. (A) The antigen is shown in purple with the glycan groups in red, and the chosen target epitope is shown in gold at the centre (the ACE2 binding site in the RBD of SARS-CoV-2, PDB ID 6VSB). The bubbles on the left show two matching antigen-like fragments (salmon) in the context of their native structure structurally superimposed to the query epitope fragments (gold) used to identify them. The CDR-like fragments (blue and green) interacting with these antigen-like fragments, and therefore predicted to bind the epitope, are also shown above it in the centre. The antigen-like fragments in the bubbles are non-contiguous as they were identified with the surface-patch fragmentation mode (see Methods) by employing as query those epitope fragments corresponding to solventexposed sidechains. The sequence identity between the query epitope fragment (gold) and matching antigen-like fragment (salmon) is shown as an alignment next to the bubbles. (B) As the two CDR-like fragments have an overlapping part with compatible backbone conformation and similar sidechain orientation they are merged together to form a single designed CDR candidate. (C) The sequence of the CDR candidate is further optimised (see Methods) by substituting those residues in contact with amino acids that differ between the target epitope and the matching antigen-like fragments. (D) The optimised CDR sequence is then grafted onto a single-domain antibody scaffold. The example presented in this figure is the CDR3 of DesAb-RBD-C1 (Table 1). The model in this panel was generated with the ABodyBuilder web server (22).
Finally, the researchers combined these to generate longer CDR loops or multiple candidates and optimized them to yield the final designs.
Designs and biophysical characterization
The researchers generated the six single-domain antibodies against the three different antigens: 1) the SARS-CoV-2 spike protein’s receptor-binding domain (RBD), 2) three human serum albumin (HSA), and one pancreatic bovine trypsin.
They observed competitive binding of the receptors, which suggests that these affinity-matured versions of the antibodies may have neutralizing potential. They also reported that all the designed single-domain antibodies exhibited excellent biophysical properties.
Conclusion
This study has demonstrated a fragment-based method for the combinatorial design of antibody binding loops and their grafting onto antibody scaffolds.
The highlight of this study is that the observed results are achieved without the need for screening a large number of designs experimentally. Instead, the researchers have pre-selected those designed CDRs in silico that appeared most promising according to the metrics implemented: proxies for the predicted binding, sidechain complementarity, and predictions of solubility.
Without involving approximations to calculate the interaction of free energies, this combinatorial approach based on the sampling of conformational and mutational space is faster than other approaches.
This said, the researchers have pointed to an intrinsic limitation of this strategy. Namely, that the applicability of this approach depends on the availability of suitable CDR-like fragments in the databases. Nonetheless, the growing number of available protein structures in public databases makes the procedure generally applicable, as for most epitopes, one obtains several candidate CDRs to choose from, the researchers argued.
We have exploited recent advances in protein-folding predictions and ab initio structural modelling to show that our design pipeline yields similar results when running on experimental structures or computer-generated models, even when these do not reach atomistic accuracy.”
Leveraging such computational biotechnology work as described in this study will enable scientists to rapidly generate lead antibodies soon after the release of a pathogen genome or from the identification of a novel disease-relevant epitope – whatever the target may.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Mauricio Aguilar Rangel, Alice Bedwell, Elisa Costanzi, Stefano Ricagno, Judith Frydman, Michele Vendruscolo, Pietro Sormanni. Fragment-based computational design of antibodies targeting structured epitopes. bioRxiv 2021.03.02.433360; doi: https://doi.org/10.1101/2021.03.02.433360, https://www.biorxiv.org/content/10.1101/2021.03.02.433360v1
- Peer reviewed and published scientific report.
Aguilar Rangel, Mauricio, Alice Bedwell, Elisa Costanzi, Ross J. Taylor, Rosaria Russo, Gonçalo J. L. Bernardes, Stefano Ricagno, Judith Frydman, Michele Vendruscolo, and Pietro Sormanni. 2022. “Fragment-Based Computational Design of Antibodies Targeting Structured Epitopes.” Science Advances 8 (45). https://doi.org/10.1126/sciadv.abp9540. https://www.science.org/doi/10.1126/sciadv.abp9540.