The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes the coronavirus disease (COVID-19), continues to spread globally.
With this spread, new variants and strains have emerged, posing a threat to the recently developed and distributed SARS-CoV-2 vaccines' efficacy.
Researchers at the Department of Integrative Biomedical Sciences, University of Cape Town, South Africa, found evidence of a significant change in the selective forces acting on immunologically important SARS-CoV-2 genes, such as N and S. These likely coincided with the emergence of the 501Y lineages.
In the study, published in the pre-print server medRxiv*, the team examined patterns of mutations that emerged in the SARS-CoV-2 genome during the pandemic.
SARS-CoV-2 mutation
Between December 2019 and October 2020, the virus's evolution worldwide involved a highly susceptible new host population. A mutation D614G (Asp614-to-Gly) in the viral spike protein hastened the spread of the virus.
From there, few mutations were epidemiologically significant without impacting SARS-CoV-2's pathogenesis. However, these mutations were characterized by a mutational pattern of slow and selectively-neutral random genetic drift.
Since October 2020, SARS-CoV-2 has mutation several times. Currently, three variants are actively spreading – the U.K. variant called B.1.1.7 with many mutations in the fall of 2020, the South African variant called B.1.351, and the Brazil variant called P.1.
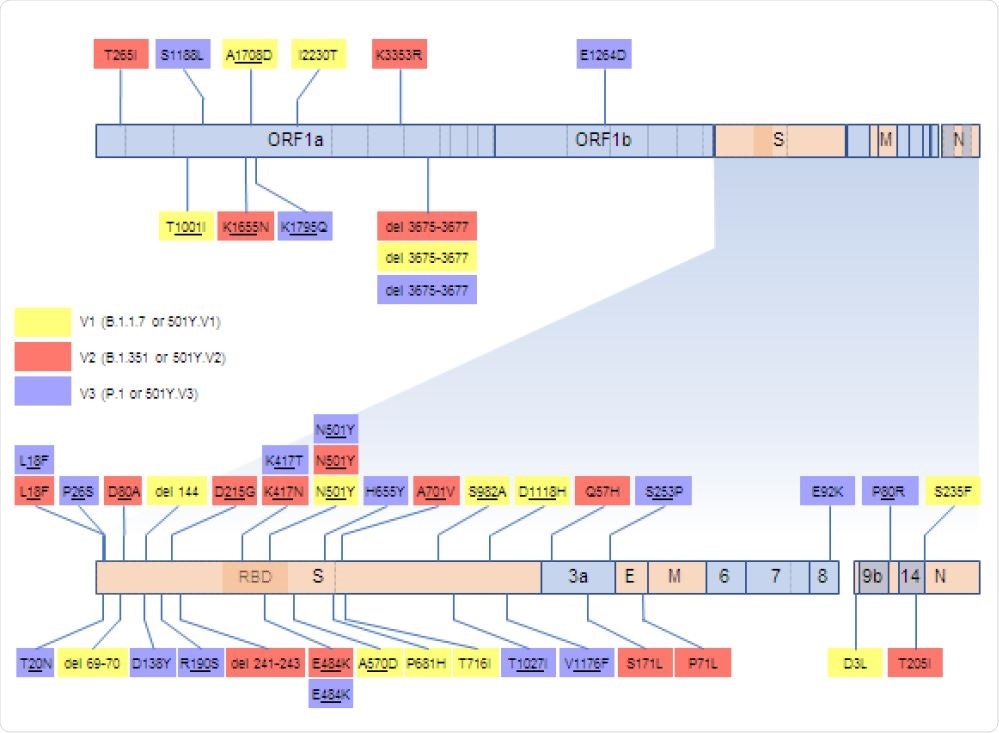
SARS-CoV-2 genome map indicating the locations and encoded amino acid changes of what we considered here to be signature mutations of V1, V2 and V3 sequences. Genes represented with light blue blocks encode non-structural proteins and genes in orange encode structural proteins:S encodes the spike protein, E the envelope protein, M the matrix protein, and N the nucleocapsid protein. Within the S-gene, the receptor binding domain (RBD) is indicated by a darker shade and the site where the S protein is cleaved into two subunits during priming for receptor binding and cell entry is indicated by a dotted vertical line.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The U.K. variant spreads faster and easier than other variants. In January 2021, scientists said that the variant might be linked to an increased risk of death than other variant viruses. It has spread to many countries worldwide.
The B.1.351 variant is known to be resistant to the effects of vaccines and therapies against COVID-19. Meanwhile, the P.1 variant emerged in travelers from Brazil in early January. This variant contains a set of additional mutations that may affect its ability to be recognized by antibodies.
In the study, the B.1.17 or 501Y.V1 variant is referred to as V1, B.1.351 or 501Y.V2 variant is V2, and P.1 or 501Y.V3 variant is referred to as V3.
To date, studies suggest that antibodies produced through vaccination with approved vaccines recognize these variants, but further investigation is ongoing.
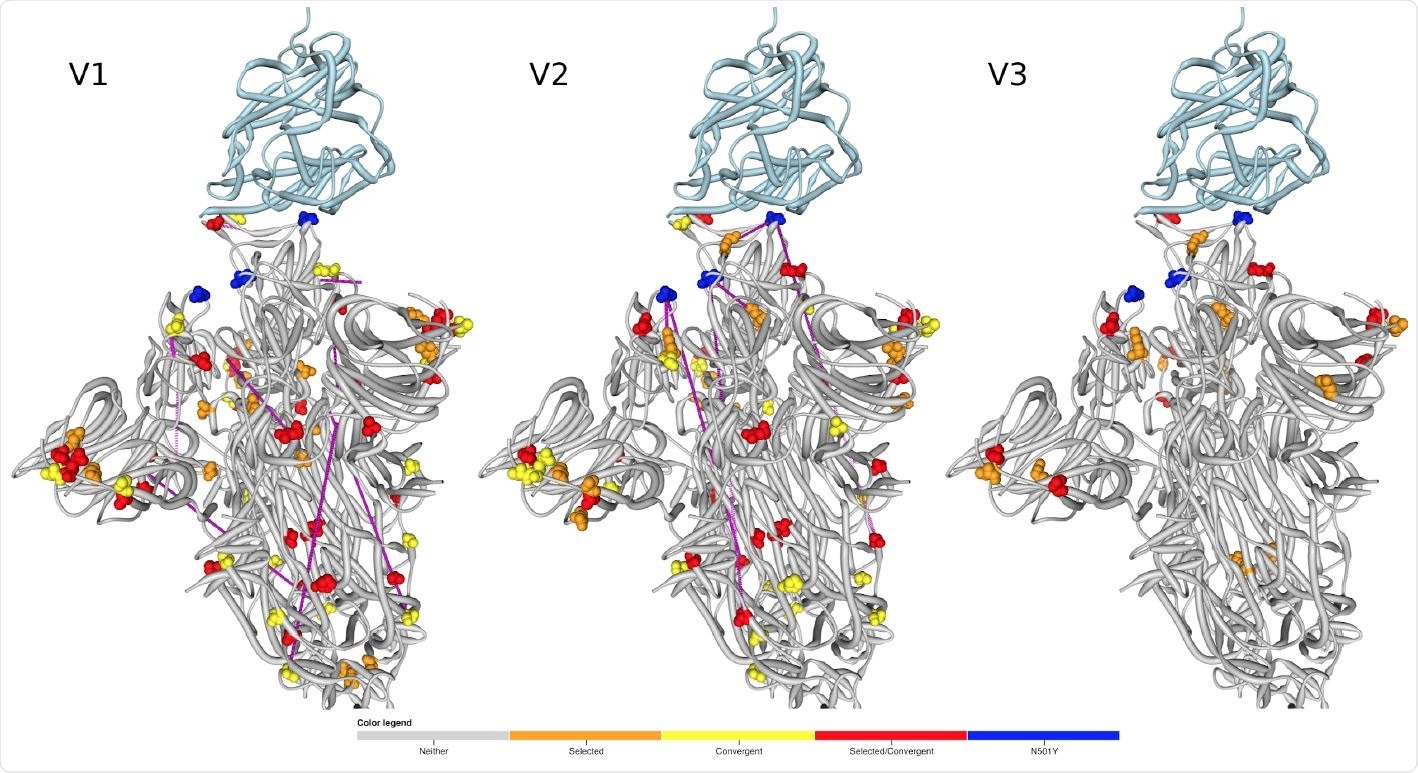
Locations of amino acids encoded by codons that are evolving under positive selection and/or encode convergent amino acid changes between the lineages mapped to the 3D structure of Spike (PDB 7DF4 structure; 47). The human ACE2 receptor is shown in light blue. Signature mutations are not represented unless they are inferred to be under positive selection. Pairs of sites that are detectably coevolving within the different lineages are linked by purple lines.
The study
The study aimed to determine the evolutionary capacity of SARS-CoV-2 to adapt to both rising population immunity and infection control measures like social distancing and vaccination.
The researchers examined patterns of mutations that emerged in SARS-CoV-2 genomes during the pandemic.
The team used a suite of phylogenetics-based natural selection analysis techniques to examine positive selection patterns within the three lineages' protein-coding sequences.
The study findings showed the emergence of the 501Y lineages concurred with a substantial global change in positive selection signals. This means a general shift in the selective environment within which SARS-CoV-2 is evolving.
They also found a significant shift in the selective forces acting on SARS-CoV-2 genes. Further, the team revealed that the adaptive evolution of the 501Y lineages unites between lineages. This study highlights the essence of surveillance on how 501Y lineage members evolve similar stages to ensure their survival and persistence.
The sudden emergence of the 501Y lineages
The team explained that in the first months of the pandemic, there were no arising mutations. This was the languid pace of the viral evolution. The pandemic's start was like a fitness peak for the virus concerning its capacity to infect and transmit between humans.
Since October 2020, however, the sudden emergence of the 501Y lineages has led to skyrocketing cases worldwide. The V1, V2, and V3 caused the virus's faster transmission, which has now reached 192 countries and regions.
"Given the numbers of infections that had occurred by October, all of these individual mutations, and even all of the potentially epistatically interacting pairs of these mutations, would have arisen independently," the researchers explained.
To date, the number of cases has reached over 117 million and 2.59 million deaths. The United States reports the highest number of infections, topping 29 million. The other countries with skyrocketing cases include India, with 11.22 million cases; Brazil, with 11 million cases, Russia, with 4.28 million cases, and the U.K., with 4.23 million cases.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Source:
Journal references:
- Preliminary scientific report.
Martin, D., Weaver, S., Tegally, H., et al. (2021). The emergence and ongoing convergent evolution of the N501Y lineages coincides with a major global shift in the SARS-CoV-2 selective landscape. medRxiv. https://www.medrxiv.org/content/10.1101/2021.02.23.21252268v1
- Peer reviewed and published scientific report.
Martin, Darren P., Steven Weaver, Houriiyah Tegally, James Emmanuel San, Stephen D. Shank, Eduan Wilkinson, Alexander G. Lucaci, et al. 2021. “The Emergence and Ongoing Convergent Evolution of the SARS-CoV-2 N501Y Lineages.” Cell 184 (20): 5189-5200.e7. https://doi.org/10.1016/j.cell.2021.09.003. https://www.cell.com/cell/fulltext/S0092-8674(21)01050-3?.