Coronavirus disease 2019 (COVID-19), caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has claimed over 2.6 million lives worldwide. Despite significant gains in scientific research since the onset of the pandemic in late-2019, much remains to be known about this virus and its biology.
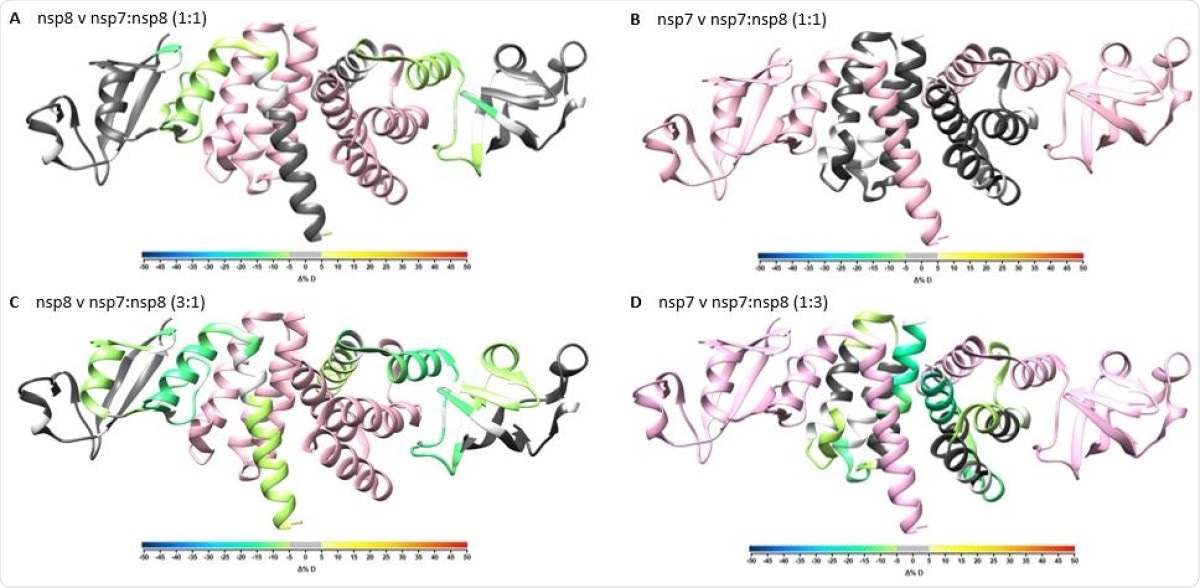
Overlay of differential HDX-MS perturbation values onto PDB:6YHU. Perturbation values for nsp7 vs nsp7:nsp8 1:1 (A), nsp8 vs nsp7:nsp8 1:1 (B), nsp7 vs nsp7:nsp8 1:3 (C), and nsp8 vs nsp7:nsp8 3:1 (D) colored according to change in percent deuterium levels shown in color bar and respective partner protein colored in pink. Residues not observed by HDX-MS are colored in white.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The RNA polymerase complex
Currently, it is thought is that at the fundamental level, the RNA polymerase complex is formed of the non-structural protein (NSP) 12, encoding the RNA-dependent RNA-polymerase (RdRp) catalytic domain, bound to NSP7 and NSP8 as cofactors, in a ratio of 1:1:2.
The structure of NSP7-NSP8 without NSP12 is as yet unknown, but in the three-protein complex, it has been suggested to form a dimer, a linear heterotetramer or a cubic heterotetramer.
Complementary techniques
The current study used a combination of hydrogen-deuterium exchange MS (HDX-MS) and crosslinking MS (XL-MS) to explore the dynamic interactions between NSP7 and NSP8.
The study, using structural proteomics, shows a high level of architectural plasticity not seen from earlier structural studies based on X-ray crystallography or cryo-electron microscopy. The latter methods, being snapshot-like profiles of protein structure, may not completely reveal this aspect of proteins in solution.
Structural plasticity is an inherent and important part of the protein structure and functions by itself and as part of a protein complex.
HDX-MS is based on hydrogen exchange between the protein backbone and the solvent, depending on several factors: the amino acid sequence itself, the folding of the protein and changes in the hydrogen bonding network.
It can help reveal the secondary structure of a protein, since both α-helices and β-sheets in the protein structure are held together by hydrogen bonds. This technique shows that NSP7 and NSP8 have a heterotetrameric structure. However, it also suggests that NSP8 may not have a helical structure when not in a complex.
The use of HDX-MS also helped confirm the NSP7-NSP8 binding sites, as well as validating the direct interactions between these proteins.
NSP7 and NSP8
When examined by XL-MS, NSP7 appeared to be a linear protein with only dead-end crosslinks. It appeared to lack the ability to crosslink with itself, or to form significant interactions between its side-chains – a process called homomultimerization. In the presence of NSP8, however, crosslinking to NSP8 occurs at the dead-end crosslink sites, forming four inter-NSP7-NSP8 crosslinks.
NSP8 crosslinks with itself at 25 sites, but some of these, near the Lys79 residue, are involved in the crosslinking with NSP7, favoring the latter interaction when NSP7 is present.
Changing the molar ratio
Since many earlier cryo-EM studies have shown that the ratio of NSP12:NSP7:NSP8 is 1:1:2, the analysis was repeated using XL-MS at a molar ratio of NSP7:NSP8 at 1:2. This brought to light four new crosslinks between them. Most of the crosslinks detected earlier remained unchanged, but a few were increased at this ratio.
The presence of these enriched intra-nsp8 crosslinks suggests that nsp8 interacts with nsp7 and nsp8 simultaneously.”
When both the techniques are overlaid, their complementary roles are brought out. The crosslinks are shown to occur at the same regions shown to be protected from hydrogen isotopic exchange from the solvent. The highest magnitude of crosslinking and of protection are those which contain both NSP Lys79 and Lys96, within H1NSP8, as expected from the X-ray structure.
Thus, the H1NSP8 site stabilizes both the dimer and the heterotetramer interface, with additional contacts being likely outside these helical bundle interactions.
Three-dimensional structure
These results can help understand the three-dimensional structure of the complex. With all the NSP7-NSP8 crosslinks being mapped to one of the interfaces in the linear heterotetramer structure, it is likely that this better represents the structure of the complex in solution.
Using the published NSP7-NSP8 linear heterotetramer conformation, 10 out of 11 crosslinks were mapped (except for one that was outside the truncated sequence used in this study). All the crosslinks were traced to the dimer or the heterotetramer interface, but only two to both.
These two are within the two helices that are suggested to form both the dimer and the heterotetramer interfaces.
The mapping of these crosslinks also allows the RTC complex to be better understood.
The results presented here suggest that CoV-2 does not assemble into a hexadecameric structure, and thus would require RNA to be able to bind the nsp7:nsp8 dimer or heterotetramer to support primer de novo synthesis.”
The researchers also found, from their mapping of the crosslinks onto the structure of the replicating polymerase, that the NSP7-NSP8 binding does not undergo change when the complex binds to NSP12. The second NSP8 may be prebound to NSP12 as it fails to make any contact with NSP7.
Conclusion
The use of these complementary techniques allows protein backbone changes to be understood, as well as the side chain structure and reactivity. This provides a better understanding of the three-dimensional structure of the protein to understand how the protein structure shapes its function.
In the current study, the crystal structures suggest three structural possibilities, which are narrowed down to one – the linear heterotetramer – which is not disturbed by the presence of RNA.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources