As the coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), continues to unfold, detecting early warning signs of new or potential outbreaks are crucial.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The UK variant
The UK variant (B.1.1.7 ) is a novel variant that has been spreading rapidly across the UK, especially in London, the South-east and the East of England. First identified in November 2020, it was then traced back to Kent, in South-East England, probably having emerged there in September.
This variant is characterized by a set of mutations, many of which affect the spike and are thought to affect the virus's biology.
These mutations include N501Y on the receptor-binding domain (RBD) of the virus, P681H near the furin cleavage site, and deletion of amino acids 69-70. These are associated with increased binding affinity to the host cell receptor – angiotensin-converting enzyme 2 (ACE2) receptor – as well as viral infectivity and immune evasion.
This striking association of the novel mutations in this variant with the potential for increased transmission, immune evasion and virulence of the virus has led to its being named SARS-CoV-2 Variant of Concern (VOC) 202012/01 by Public Health England (PHE).
Deletion of amino acids 69/70 is associated with S-gene target failure (SGTF) in polymerase chain reaction (PCR) diagnostic testing for the virus when the diagnosis has been confirmed by the recognition of other gene targets. Such SGTF is used to arrive at the probable fraction of confirmed COVID-19 cases caused by the UK variant.
Increasing prevalence
Both whole-genome sequencing and SGTF data have shown that the prevalence of this variant has increased rapidly over time, such that 94% of sequenced clinical samples from England by the end of January show the presence of this variant. Epidemiological data shows that this lineage may be more transmissible than the earlier D614G strain.
The reproduction number of this lineage also appears to have gone up by 0.4 and 0.7, relative to the earlier strains.
The UK variant was followed in quick succession by the South African and Brazil variants, both of which have their own unique and important mutations that further affect immune evasion properties. In fact, the mutation E484K, which confers partial resistance to neutralizing antibodies, is present in some viruses of the B.1.1.7 lineage, possibly further increasing its danger to public health.
Environmental surveillance
Environmental surveillance (ES) is thus indicated as a method suitable for monitoring the spread of the virus worldwide and rapidly picking up genomic changes as well as epidemiological departures from the normal, indicating a change in viral biology.
Not only has ES proved its worth in multiple settings for this purpose, but it can also provide a precise idea of the prevalence of VOCs in the population. Earlier ES based on sewage examination from samples collected in London during the first wave of the pandemic showed a close correspondence between the levels of viral RNA and the COVID-19 caseload.
Next-generation sequencing (NGS) of DNA amplicons from sewage allowed changes in the most prevalent variants to be tracked, from the earlier strains to the almost completely dominant D614G strain by May 2020. This strain appears to be far more transmissible than the ancestral strain.
Increased sewage viral RNA load precedes outbreaks
The current study aims to detect the presence of SARS-CoV-2 in London sewage samples using PCR against two gene targets, with a semi-quantitative nested PCR (nPCR) assay in preference to the quantitative PCR tests, that may be less sensitive and consistent.
The researchers also developed new nPCR reactions to recognize DNA amplicons, as well as to examine them using NGS, and thus to pick up and quantify important mutations that distinguish the three main VOCs from the ancestral lineages and from each other.
While sewage collected in January 2020 was negative for viral RNA, and a February sample contained low levels of the virus, viral RNA shot up in March and April, known for the first wave. In May, the national lockdown was associated with a steep fall in viral RNA levels in sewage, which was sustained for four months.
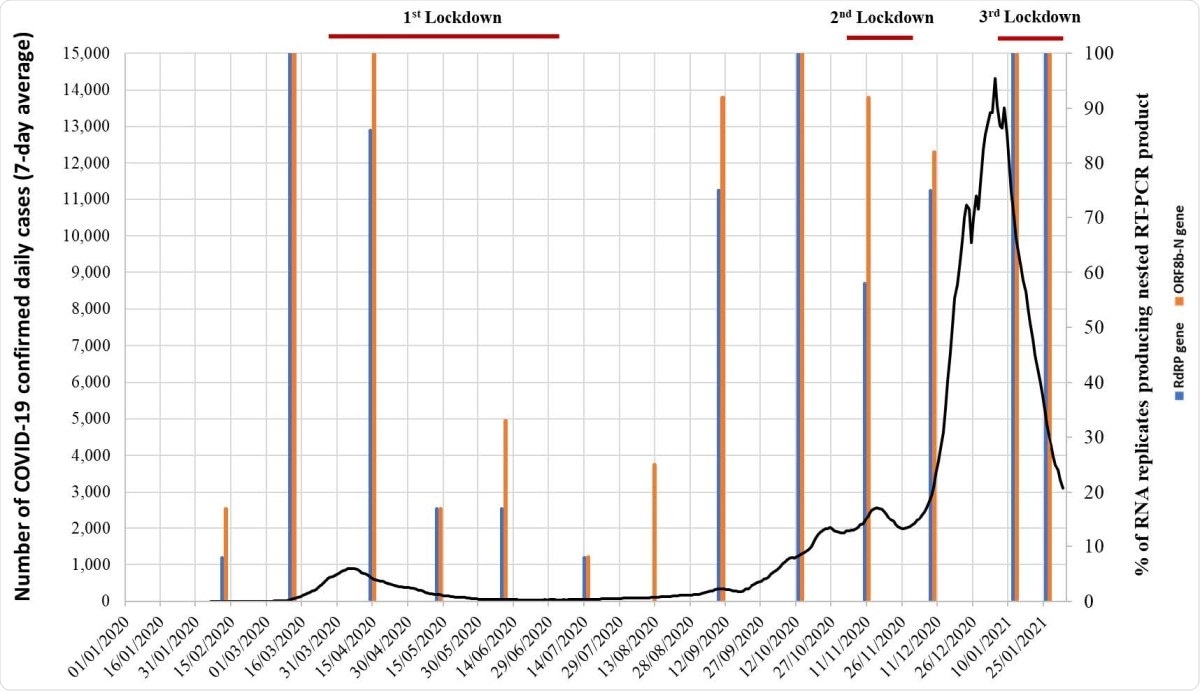
Detection of SARS SARS-CoVCoV-2 RNA in sewage samples collected in London between 14 14th January 2020 and 26 26th January 2021. The % of RNA replicates producing positive results in nPCR A (blue) and nPCR B (orange) reactions targeting RdRP and ORF8b genomic regions, respectively, are shown as vertical columns. The number of daily confirmed COVID COVID-19 cases in London are represented with a black line (7 7-day rolling average). Nationwide lockdown periods are indicated as horizontal red lines. Source for COVID COVID-19 cases data: https://coronavirus.data.gov.uk/
The next wave began in September with increasing viral RNA levels.
The second lockdown period lasting from 5th November to 2nd December appear to only have had a limited effect in reducing virus transmission which, together with the emergence of the apparently highly transmissible B.1.1.7 variant, may explain the high levels of viral RNA found in sewage and the high number of SARS-CoV-2 infections reported in England in December 2020 and January 2021."
In fact, in the week ending January 2, one in 30 and one in 50 people in London and England were thought to have acquired the infection.
NGS of the samples showed there was an increase in the mutations that distinguish the UK lineage from the ancestral strains from November 2020 to January 2021. In quantitative terms, the mutations increased from about 6.5-9% to 95-99% over this period.
The resulting estimates of the prevalence of the UK variant agreed well with SGTF and whole-genome sequencing data.
The frequency rates of B.1.1.7 mutations found in viral RNAs from sewage samples and from sequences derived from clinical samples showed excellent concordance, but this was not found with the mutations unique to the other two VOCs.
What are the implications?
The very high agreement between the changes in viral RNA load in sewage samples and clinical samples from London shows the feasibility and reliability of using wastewater as a means of ES during the COVID-19 pandemic.
ES has a potential advantage for the early detection of VOCs versus clinical surveillance as it provides an immediate snapshot of virus transmission events representing thousands/millions of people, including asymptomatic infections."
Using methods adopted during this study, sequencing results could be available within a week of the sewage samples reaching the laboratory. This snapshot of the virus in circulation in a large population is thus superior in pace to the detection of transmission patterns by the sequencing of clinical specimens, with the inevitable delay from infection to symptom onset and then to testing, and finally to the reporting of the test results.
The detection of B.1.1.7 mutations shows the possibility of using this tool to identify other mutations, notably those of the South African and Brazil variants, if they emerge, ahead of their clinical appearance. This could also help to develop updated vaccines based on the strains that are in circulation at that point.
Clinical surveillance demands rapid and extensive PCR testing, with a very large whole-genome viral sequencing program, which is confined to very few countries at present. In this situation, ES could be a boon to predict outbreaks and identify VOCs.
Rather than ES on sewage samples from wastewater treatment sites serving large populations, community-based ES would help detect VOCs more sensitively.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Wilton, T. et al. (2021) Rapid increase of SARS-CoV-2 variant B.1.1.7 detected in sewage samples from England between October 2020 and January 2021. medRxiv preprint. doi: https://doi.org/10.1101/2021.03.03.21252867, https://www.medrxiv.org/content/10.1101/2021.03.03.21252867v1
- Peer reviewed and published scientific report.
Wilton, Thomas, Erika Bujaki, Dimitra Klapsa, Manasi Majumdar, Maria Zambon, Martin Fritzsche, Ryan Mate, and Javier Martin. 2021. “Rapid Increase of SARS-CoV-2 Variant B.1.1.7 Detected in Sewage Samples from England between October 2020 and January 2021.” Edited by Charles R. Langelier. MSystems 6 (3). https://doi.org/10.1128/msystems.00353-21. https://journals.asm.org/doi/10.1128/mSystems.00353-21.