In a further development of the collaboration between QCMD and RNAssist, QCMD will use the universal biological sample stabilization virusPHIX-P9™ as the matrix within its forthcoming SARS-CoV-2 antigen EQA pilot scheme.
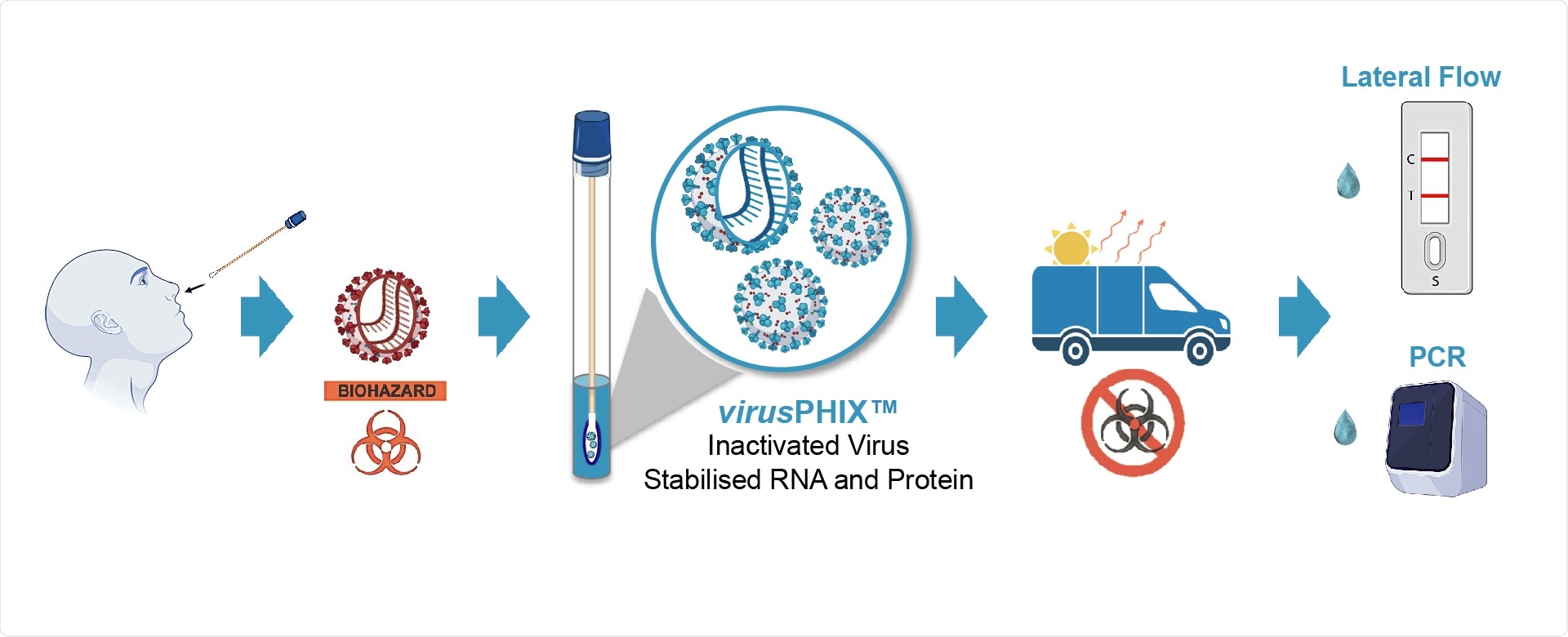
Image Credit: Rapid Labs
Studies have proven that virusPHIX-P9™ is both capable of inactivating SARS-CoV-2 as well as stabilizing clinical samples at ambient temperatures. Extensive testing at QCMD has shown that virusPHIX-P9™ is compatible with a variety of SARS-CoV-2 antigenic tests including a wide range of Lateral Flow tests (LFT).
The QCMD SARS-CoV-2 Antigen EQA Pilot Scheme will consist of both positive and negative whole inactivated SARS-CoV-2 in virusPHIX-P9™ and will allow the performance assessment of applied clinical laboratory antigenic tests as well as rapid antigenic tests such as lateral flow assays used within public health practice. Further information can be obtained via the QCMD website.
RNAssist and Rapid Labs have developed virusPHIX-P9™ as a universal Virus Transport Media (VTM) specifically for the safe transport of clinical swab and saliva samples to testing labs as well as for EQAs and molecular controls. virusPHIX-P9™ is the only VTM that inactivates virus and stabilizes nucleic acid, which can also be used directly for Lateral Flow and PCR tests. More information can be found https://www.rapidlabs.co.uk/.
This license agreement strengthens the existing relationship between RNAssist/Rapid Labs and QCMD in the area of sample stabilization technologies, it shows our focus on establishing partnerships to maximize the potential of our stabilization technologies for a wide array of clinical and research applications.”
Andrew Goldsborough, the CEO of RNAssist
“The use of virusPHIX-P9™ in QCMD’s latest SARS-CoV-2 antigen EQA pilot scheme will allow us to further evaluate the full potential of this novel and innovative reagent. Stated Paul Wallace, QCMD Executive.
virusPHIX-P9™ and all RNAssist reagents are manufactured and distributed globally by Rapid Labs Ltd. For more information please contact us at [email protected].