Researchers have found a new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant in California, USA. This variant is more transmissible than circulating strains but slightly less transmissible than the new N501Y variant and moderately resistant to antibodies in convalescent sera and mRNA vaccines.
SARS-CoV-2 has a slower mutation rate compared to other RNA viruses. However, because of the coronavirus disease 2019 (COVID-19) pandemic and high transmissibility, the virus has acquired several mutations, with the D614G currently being the predominant variant globally.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Emerging mutations in the virus may affect transmission, infectivity, and our immunity to them. This is why some have been termed variants of concern (VOCs). Three VOCs emerged starting late-2020, having several mutations in the genome, including the receptor-binding domain (RBD) of the spike protein.
The variant from the United Kingdom (UK), B.1.1.7, has 17 mutations, including N501Y, which makes it more transmissible than other circulating variants. This variant has now become the predominant strain in the UK and Europe.
The two other VOCs detected first in South Africa, and Brazil have the E484K and K417N/K417T mutations in addition to E484K. Studies have suggested that the E484K mutation may be resistant to antibody neutralization, reducing vaccine efficacy.
Another variant was reported in January 2021 in California with the L452R mutation. Researchers report the spread of this variant in California in a paper published in the medRxiv* preprint server.
Characterizing new virus variant
The researchers sequenced 2,172 viral genomes in 44 California counties from samples that tested positive for SARS-CoV-2. The new variant, called 20C/L452R or B.1.427/B.1.429, was seen in about 21% of the genomes. This genome grew from 0% in September 2020 to more than 50% of the sequenced genomes in January 2021, a rapid increase in the number of cases compared to the other circulating variants.
Further analysis revealed two different lineages in clade 20C, B.1.427 and B.1.429. Both have three mutations in the spike protein (S13I, W152C, and L452R), another coding mutation in the orf1b protein, and two non-coding mutations. Four more mutations were specific to B.1.427, and three others were specific to B.1.429. These lineages likely diverged in June-July 2020.
Although the sudden appearance of several new mutations in a new virus variant is not unexpected, the evolutionary mechanism remains unknown. The divergence could have occurred by accelerated quasispecies evolution in patients infected for a long time.
The median PCR cycle threshold value (Ct) associated with the new variants was much lower than that of the non-variants. The viral RNA in swab samples is about 2-fold higher in the new variants.
To understand how the L452R variant affects viral entry, the team used pseudoviruses with different mutations and infected 293T cells expressing the angiotensin-converting enzyme 2 (ACE2) receptor and TMPRSS2 protein and human airway lung organoids. They found viruses with the L452R mutation had up to 22.5-fold and 14.7-fold higher entry in 293T and the organoids, respectively, slightly lower than viruses with the N501Y mutation. So, one question is if the L452R variant would remain to be the predominant strain in California or if it would be replaced by the N501Y variant (B.1.1.7).
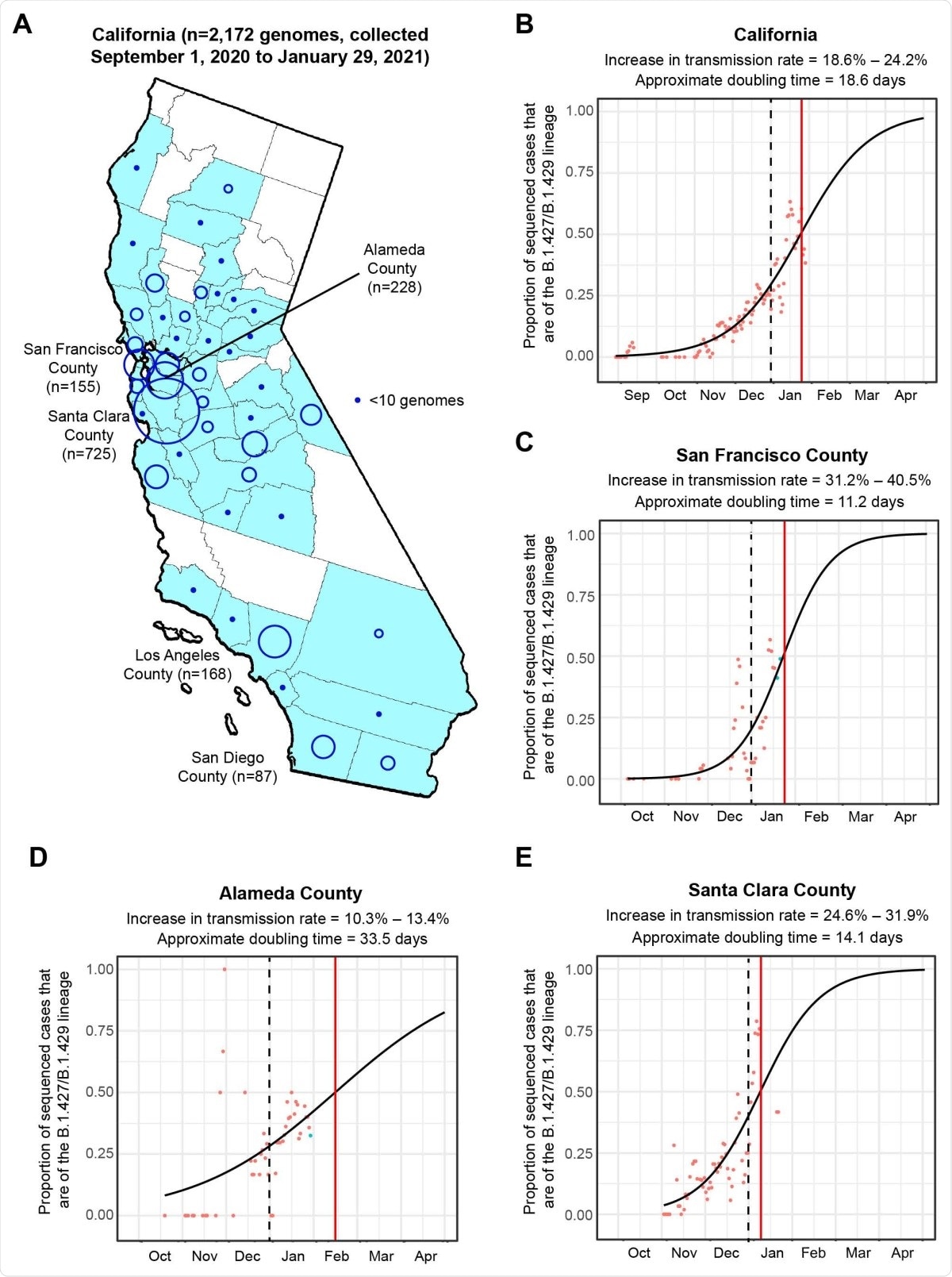
Increasing frequency of the B.1.427/B.1.429 variant 304 in California from September 1. 2020 to January 29, 2021. (A) County-level representation of the 2,172 newly sequenced SARS-CoV-2 genomes in the current study. Counties from which at least 1 genome were sequenced are colored in sky blue. The size of the circle is proportionally to the number of genomes sequenced from each county, while points designate counties for fewer than 10 genomes were sequenced. Logistic growth curves fitting the 5-day rolling average of the estimated proportion of B.1.427/B.1.429 variant cases in (B) California, (C) San Francisco County, (D) Alameda County, and (E) Santa Clara County. The predicted time when the growth curve crosses 0.5 is indicated by a vertical red line. A vertical black dotted line denotes the transition from 2020 to 2021. The increase in transmission rate is defined as the change in the relative proportion of B.1.427/B.1.429 variant cases relative to circulating non-B.1.427/B.1.429 variant lineages as estimated from the logistic growth model (Volz et al., 2020; Washington et al., 2021).
New variant moderately resistant to neutralizing antibodies
The team also tested the effect of convalescent plasma and samples from vaccinated individuals in neutralizing the variant virus. About 88% of the convalescent sera and 55% of the Pfizer or Moderna vaccine recipients showed reduced titers to the B.1.429 lineage in plaque reduction neutralization tests. Neutralization antibody response tests showed reduced neutralization antibody titers for the B.1.427 lineage. Thus, the new variants were resistant to antibodies from prior infection by about 4 to 6-fold and to vaccination by about 2-fold.
This suggests immune selection pressure could be partly responsible for the mutations. However, mRNA vaccines seem to provide robust antibodies in response to these variants selected under immune pressure.
The results suggest that the variants B.1.427 and B.1.429 require continued investigation and monitoring for potential future COVID-19 surges and reduced vaccine efficacy.
Although it is likely that these variants emerged as early as May 2020, they were not identified in the United States until July and September 2020 for the B.1.429 and B.1.427 lineages, respectively. This is probably because of poor genomic sequencing of circulating viruses.
The first public disclosure of this variant was done only in January 2021, and by then, the variants had become the predominant strains in several California counties and had spread to other states. Earlier identification could have allowed better contact tracing and timely investigation of its significance.
The results here highlight the urgent need for implementation of a robust genomic surveillance system in the US and globally to rapidly identify and monitor SARS-CoV-2 variants,” write the authors.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Deng, X. et al. (2021) Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv. https://doi.org/10.1101/2021.03.07.21252647, https://www.medrxiv.org/content/10.1101/2021.03.07.21252647v1
- Peer reviewed and published scientific report.
Deng, Xianding, Miguel A. Garcia-Knight, Mir M. Khalid, Venice Servellita, Candace Wang, Mary Kate Morris, Alicia Sotomayor-González, et al. 2021. “Transmission, Infectivity, and Neutralization of a Spike L452R SARS-CoV-2 Variant.” Cell 184 (13): 3426-3437.e8. https://doi.org/10.1016/j.cell.2021.04.025. https://www.cell.com/cell/fulltext/S0092-8674(21)00505-5?.